Introduction - 2qgnA: Difference between revisions
Choojinhsien (talk | contribs) No edit summary |
No edit summary |
||
| Line 9: | Line 9: | ||
The crystal structure of tRNA-IPT has been identified from Bacillus Halodurans (PDB code: 2QGN).With a length of 322 amino acids, it is composed of two main domains with a sulfate ion ligand embedded in its structure. Here in this study, we present data collected from different sources on the analysis of this essential enzyme. Although the structure of the human tRNA-IPT has yet to be identified, the cDNA sequence has been recently cloned. Bioinformatic tools and analysis help to compare IPT protein sequences and structures from different organisms so that its evolution and structural conservation can be studied. This allows the function of this enzyme to be determined. Bioinformatic tools have helped to determine the possible functions of tRNA-IPT there are several conserved regions that might contribute to the functional role of these enzymes. | The crystal structure of tRNA-IPT has been identified from Bacillus Halodurans (PDB code: 2QGN).With a length of 322 amino acids, it is composed of two main domains with a sulfate ion ligand embedded in its structure. Here in this study, we present data collected from different sources on the analysis of this essential enzyme. Although the structure of the human tRNA-IPT has yet to be identified, the cDNA sequence has been recently cloned. Bioinformatic tools and analysis help to compare IPT protein sequences and structures from different organisms so that its evolution and structural conservation can be studied. This allows the function of this enzyme to be determined. Bioinformatic tools have helped to determine the possible functions of tRNA-IPT there are several conserved regions that might contribute to the functional role of these enzymes. | ||
Basic Local Alignment Search Tool (BLAST) enables a researcher to compare a query sequence with a library or database of sequences and identify library sequences that resemble the query sequence above a certain threshold (Altschul, Gish et al. 1990). | |||
In order to show the evolutionary relationship between species, neighbour-joining phylogenetic tree could be constructed by ClustalX. This algorithm first did a pairwise alignment of all pairs to determine sequence similarity and then define an order of addition of sequences to alignments bases on similarity and finally generate multiple alignments progressively based on defined order (Kohli and Bachhawat 2003). | |||
A bootstrap tree with bootstrap value of 100 will be drawn based on the neighbour-joining tree. Bootstrap estimation is formulated as a two-step sampling procedure: (1) sampling of sequences from the evolutionary process and (2) resampling of the original sequence sample. The probability that a bootstrap resampling of an original sequence sample will support the true tree is found to depend on the model tree, the sequence length, and the probability that a randomly chosen nucleotide site is an informative site (Zharkikh and Li 1992). A tree figure drawing tool FigTree was used (Rambaut 2006). All bootstrap trees were demonstrated in a circular layout. | |||
[[Image:pathway.jpg|framed|none]] | [[Image:pathway.jpg|framed|none]] | ||
Revision as of 09:50, 9 June 2008
Cytokinins are first discovered in plants especially in meristematic regions and growing tissues. They function in cell division, cell differentiation and cell growth. Now, cytokinins are found also in tRNA of prokaryotes and eukaryotes. They can exist in two forms; the tRNA-free and the tRNA-bound form, through addition of adenine to tRNA or AMP acceptor molecules. Nonetheless, the presence of different tRNA species along with fully modified tRNAs does not appear to inhibit the enzyme activity, as postulated by Rosenbaum and Gefter(1972).Here, cytokinins are tRNA molecules with modified adenine bases. This modification is important as it stabilises and promotes efficient translational machinery. The enzyme responsible for the biosynthesis of these cytokinins in prokaryotes and eukaryotes is tRNA isopentenyltransferase (tRNA- IPT). This enzyme is also known as isopentenyl-diphosphate:trna isopentenyltransferase or tRNA delta(2)-isopentenylpyrophosphate transferase. In yeast, it has been known to be encoded by the MOD-5 gene, responsible for both mitochondrial and cytosolic enzyme activities(Martin and Hopper, 1994).
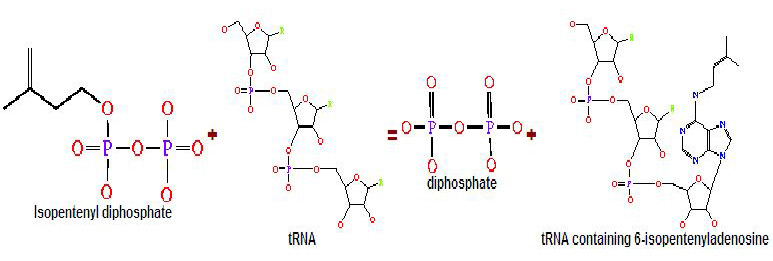
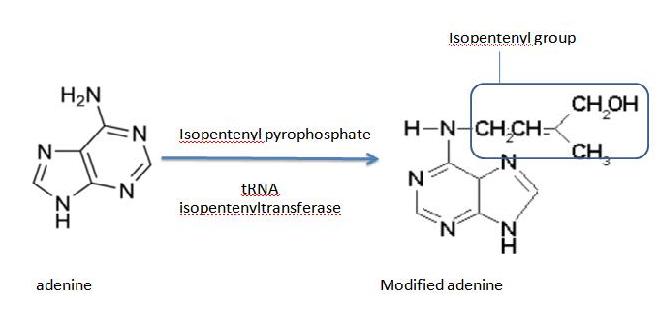
Generally, transferases are enzymes involved in tRNA metabolism, which encompasses tRNA breakdown, modification and synthesis. Essentially, this function is important in cellular processes such as protein synthesis. As for isopentenyltransferases, they transfer an isopentenyl group to the acceptor molecule. tRNA isopentenyltransferase (tRNA-IPT) as the name suggests transfers the isopentenyl group of dimethylallyl pyrophosphate to tRNA molecules. Specifically, it transfers an isopentenyl group onto adenine bases of tRNA molecules next to the anticodon which interacts with the codon encoding for uridine (Anna Golovko 2002). Modified adenine bases are commonly found at position 37 of the target tRNA.
The crystal structure of tRNA-IPT has been identified from Bacillus Halodurans (PDB code: 2QGN).With a length of 322 amino acids, it is composed of two main domains with a sulfate ion ligand embedded in its structure. Here in this study, we present data collected from different sources on the analysis of this essential enzyme. Although the structure of the human tRNA-IPT has yet to be identified, the cDNA sequence has been recently cloned. Bioinformatic tools and analysis help to compare IPT protein sequences and structures from different organisms so that its evolution and structural conservation can be studied. This allows the function of this enzyme to be determined. Bioinformatic tools have helped to determine the possible functions of tRNA-IPT there are several conserved regions that might contribute to the functional role of these enzymes.
Basic Local Alignment Search Tool (BLAST) enables a researcher to compare a query sequence with a library or database of sequences and identify library sequences that resemble the query sequence above a certain threshold (Altschul, Gish et al. 1990).
In order to show the evolutionary relationship between species, neighbour-joining phylogenetic tree could be constructed by ClustalX. This algorithm first did a pairwise alignment of all pairs to determine sequence similarity and then define an order of addition of sequences to alignments bases on similarity and finally generate multiple alignments progressively based on defined order (Kohli and Bachhawat 2003).
A bootstrap tree with bootstrap value of 100 will be drawn based on the neighbour-joining tree. Bootstrap estimation is formulated as a two-step sampling procedure: (1) sampling of sequences from the evolutionary process and (2) resampling of the original sequence sample. The probability that a bootstrap resampling of an original sequence sample will support the true tree is found to depend on the model tree, the sequence length, and the probability that a randomly chosen nucleotide site is an informative site (Zharkikh and Li 1992). A tree figure drawing tool FigTree was used (Rambaut 2006). All bootstrap trees were demonstrated in a circular layout.
Figure 1. The transfer of the isopentenyl group to tRNA molecule
Figure 2. The modification of adenine by adding the isopentenyl group