Introduction of SNAPG: Difference between revisions
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
[[Image:Snap-gamm ligands.png]] | [[Image:Snap-gamm ligands.png]] | ||
Back to [[Scientific Report of N-ethylmaleide-sensitive factor attachment protein, gamma]] | |||
Revision as of 09:51, 11 June 2007
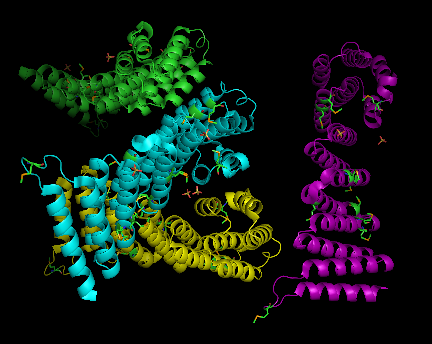
SNAPG protein structure obtained from Danio rerio organism comprises of 4 identical molecular chains (Chain A, B, C and D) interacting to one another via non-bonded contacts. Each chain is 307 amino acid residues in length and made of 69% helical secondary structure of 16 helices, 34 helix-helix interaction, 8 beta turns and 2 gamma turns[1]. Two types of molecular component were observed to formed interactions with SNAPG molecule. These molecular components were sulfanate ion(SO4) and selenomethionine (MSE)(Figure 1). The sulfanate ions interact differently to different residues of different chain, unlike MSE in which included into SNAPG amino acid sequences as modified residue (Figure 1.B,C & 2).
Back to Scientific Report of N-ethylmaleide-sensitive factor attachment protein, gamma