Next: Difference between revisions
No edit summary |
No edit summary |
||
| Line 18: | Line 18: | ||
''%id'' - percentage of identical amino acids over structurally equivalent residues. | ''%id'' - percentage of identical amino acids over structurally equivalent residues. | ||
[[Next]] | |||
Revision as of 05:04, 10 June 2008
Structural Alignment
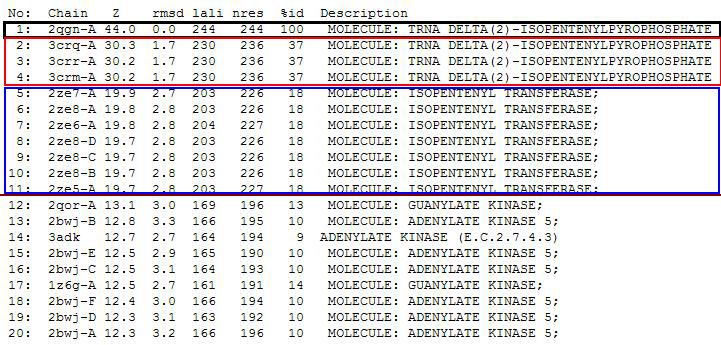
Dali Output
PDB entry code for 2qgn was loaded onto DALI server to search for structurally similar neighbours. Displayed below are the results from DALI search :-
DALI output describes the following :
Z score , the statistical significance of the similarity between protein-of-interest and other neighbourhood protines. The program optimises a weighted sum of similarities of intramolecular distances.
Root Mean Square Distance (RMSD), root-mean-square deviation of C-alpha atoms in the least-squares superimposition of the structurally equivalent C-alpha atoms. As in indicated in DALI, rmsd is not optimised and is only reported for information.
lali, the number of structurally equivalent residues.
nres, or the total number of amino acids in the hit protein.
%id - percentage of identical amino acids over structurally equivalent residues.