Phytanoyl-CoA dioxygenase Sequence and Evolution: Difference between revisions
From MDWiki
Jump to navigationJump to search
| Line 17: | Line 17: | ||
[[Image:Msa3.jpg|center|framed|'''Figure 1.3'''<Br> Multiple Sequence Alignment obtained from ClustalX]]<Br> | [[Image:Msa3.jpg|center|framed|'''Figure 1.3'''<Br> Multiple Sequence Alignment obtained from ClustalX]]<Br> | ||
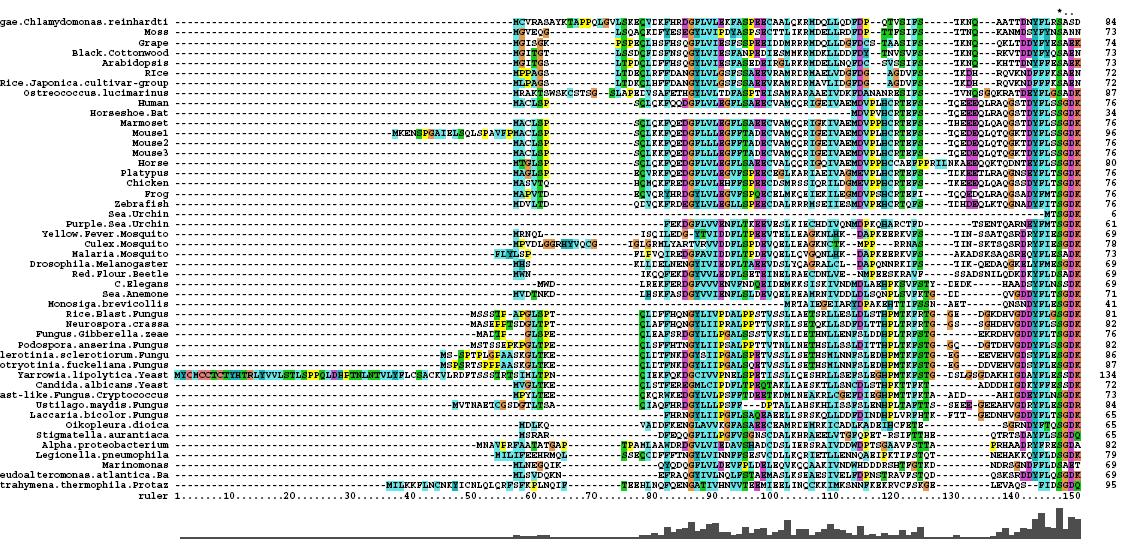
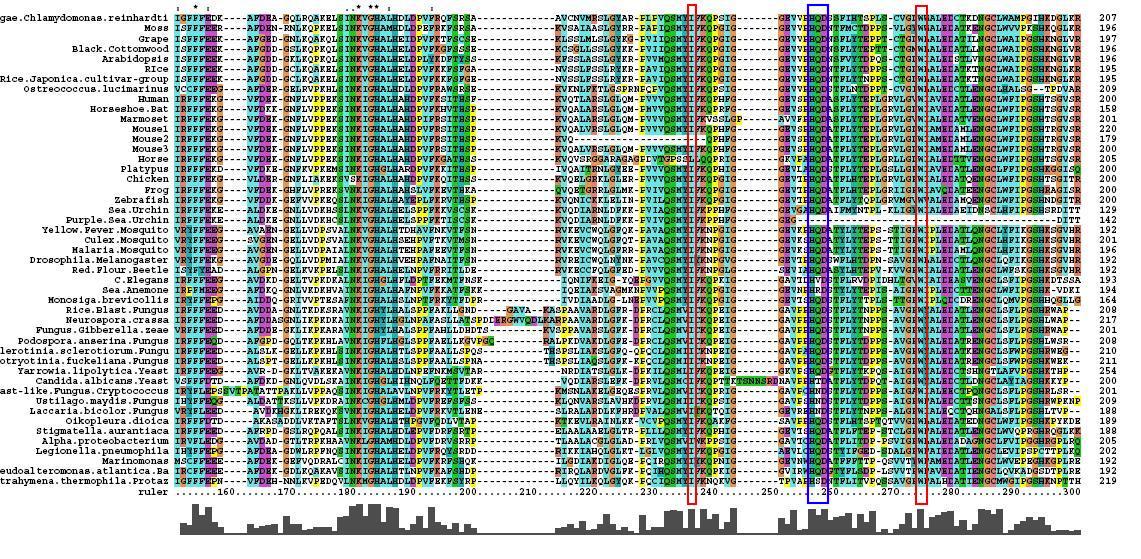
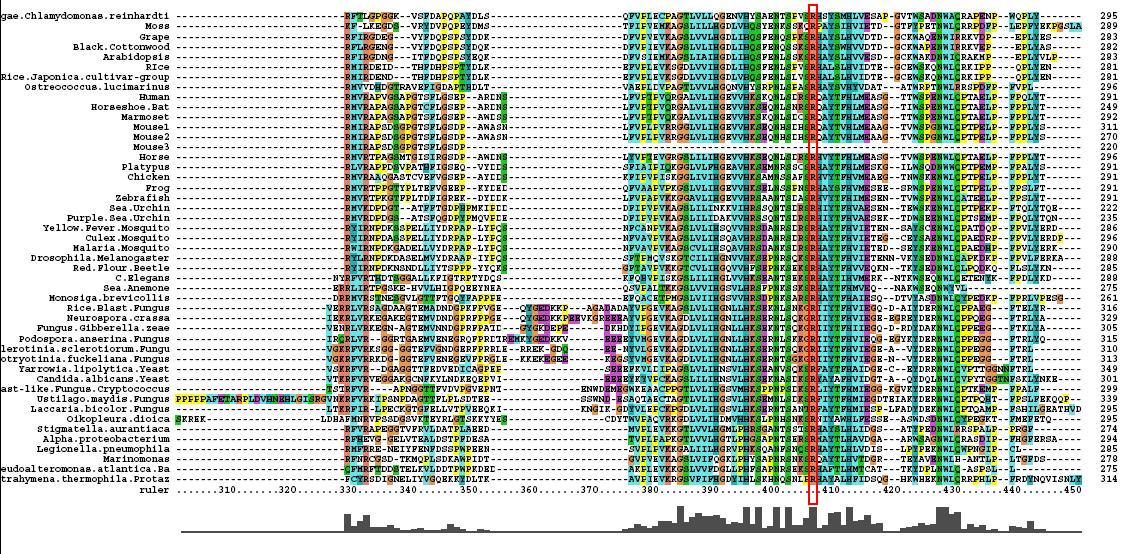
*46 analogous sequence compared using ClustalX | |||
*Multiple Sequence Alignment is of high quality and conservation | |||
*Fe2+ and 2-Oxogylterate binding sites shown | |||
== Phylogenetic Trees == | == Phylogenetic Trees == | ||
Revision as of 01:05, 10 June 2008
Fasta Sequence
>gi|134105315|pdb|2OPW|A Chain A, Crystal Structure Of Human Phytanoyl-Coa Dioxygenase Phyhd1 (Apo) MACLSPSQLQKFQQDGFLVLEGFLSAEECVAMQQRIGEIVAEMDVPLHCRTEFSTQEEEQLRAQGSTDYF LSSGDKIRFFFEKGVFDEKGNFLVPPEKSINKIGHALHAHDPVFKSITHSFKVQTLARSLGLQMPVVVQS MYIFKQPHFGGEVSPHQDASFLYTEPLGRVLGVWIAVEDATLENGCLWFIPGSHTSGVSRRMVRAPVGSA PGTSFLGSEPARDNSLFVPTPVQRGALVLIHGEVVHKSKQNLSDRSRQAYTFHLMEASGTTWSPENWLQP TAELPFPQLYT
Multiple Sequence Alignment
- 46 analogous sequence compared using ClustalX
- Multiple Sequence Alignment is of high quality and conservation
- Fe2+ and 2-Oxogylterate binding sites shown
Phylogenetic Trees
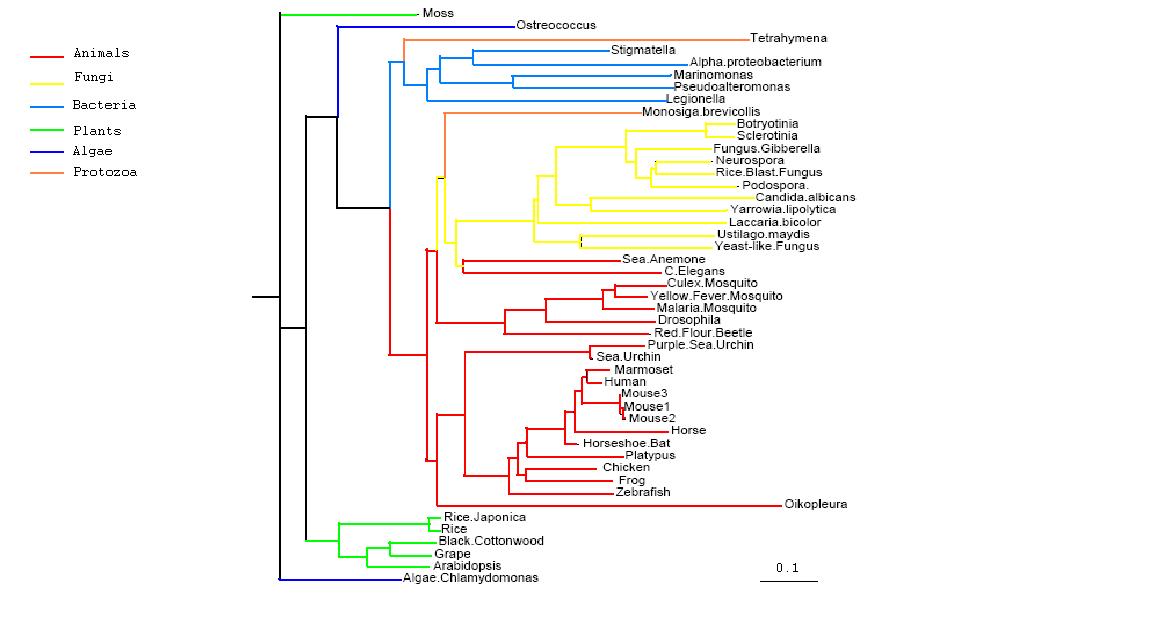
Figure 2.1
Rooted Phylogenetic Tree obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
Rooted Phylogenetic Tree obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
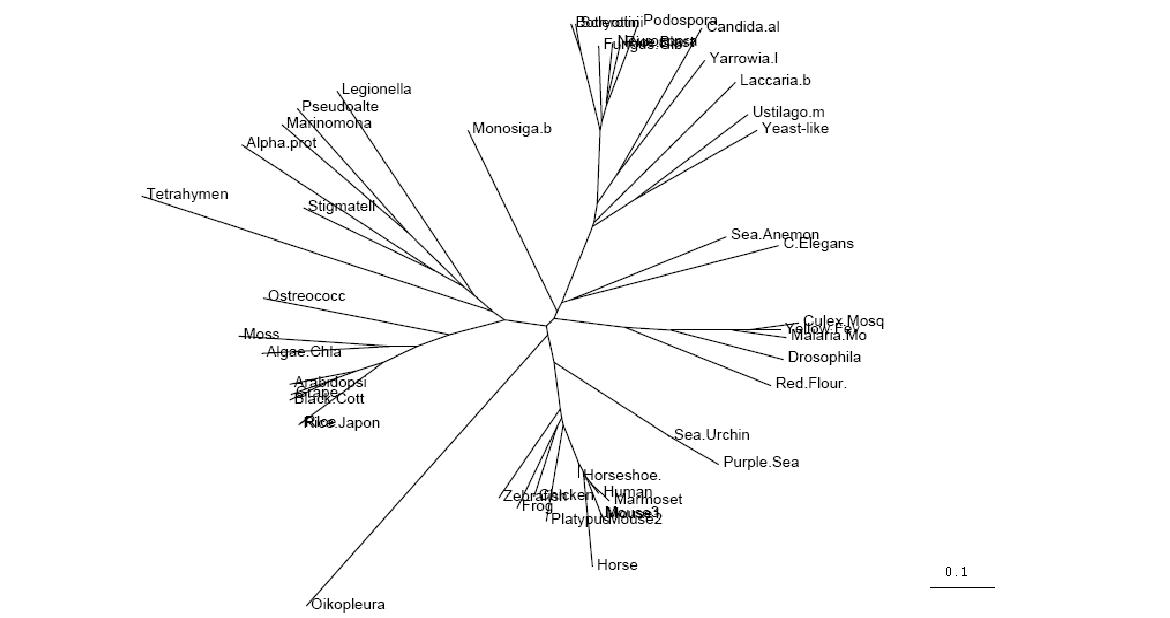
Figure 2.1
Unrooted Phylogenetic Tree obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
Unrooted Phylogenetic Tree obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
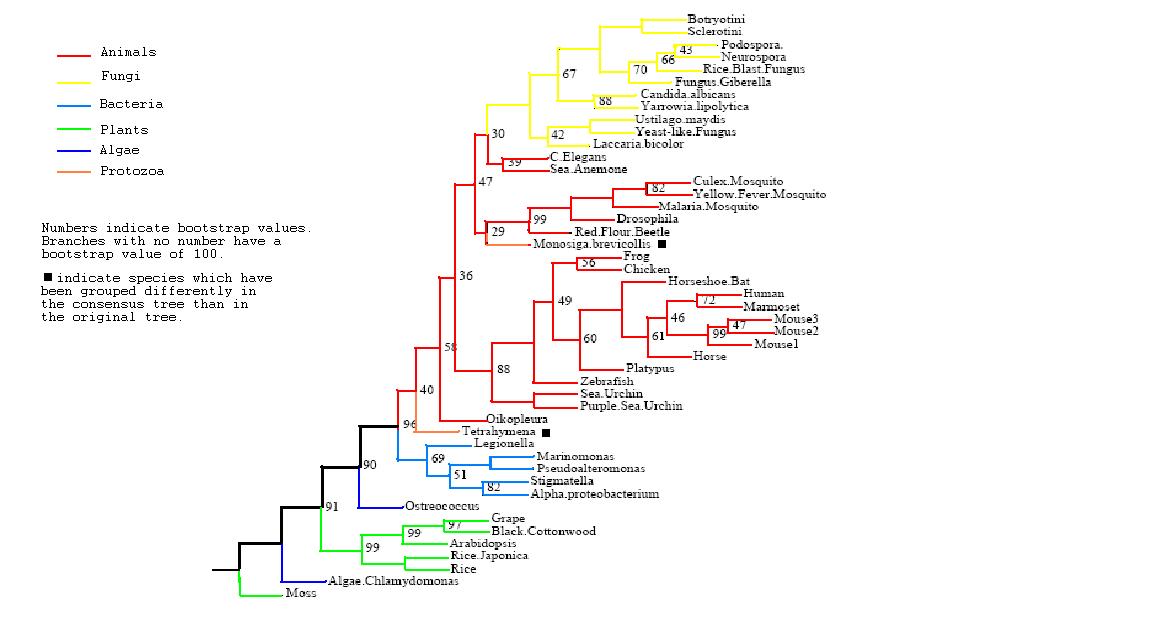
Figure 3
Rooted Consensus Tree with bootstrap values obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
Rooted Consensus Tree with bootstrap values obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/