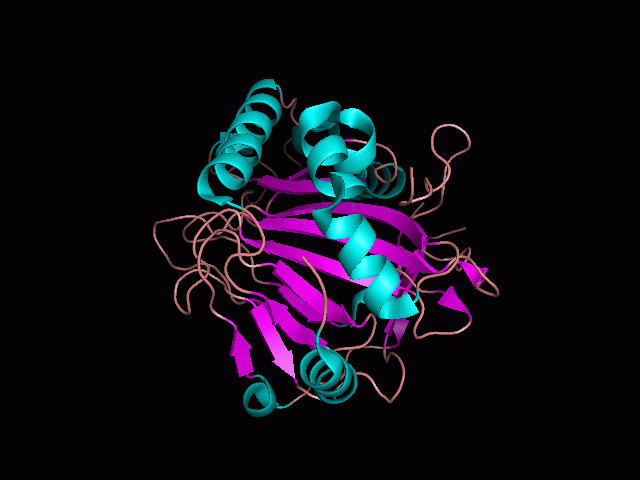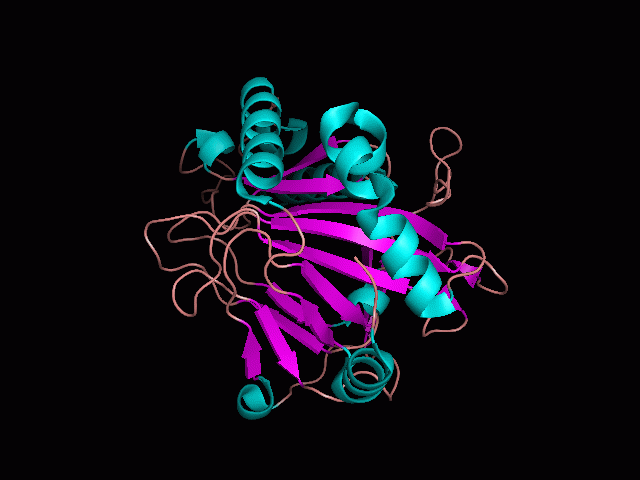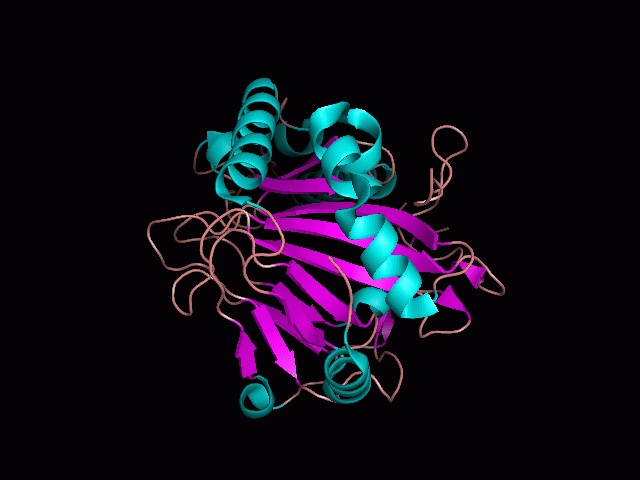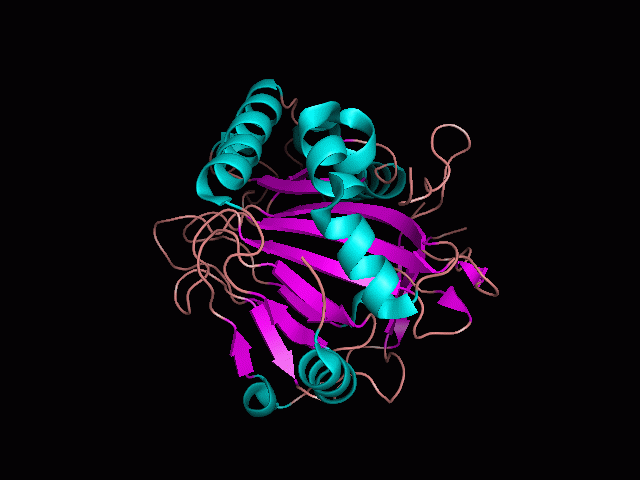
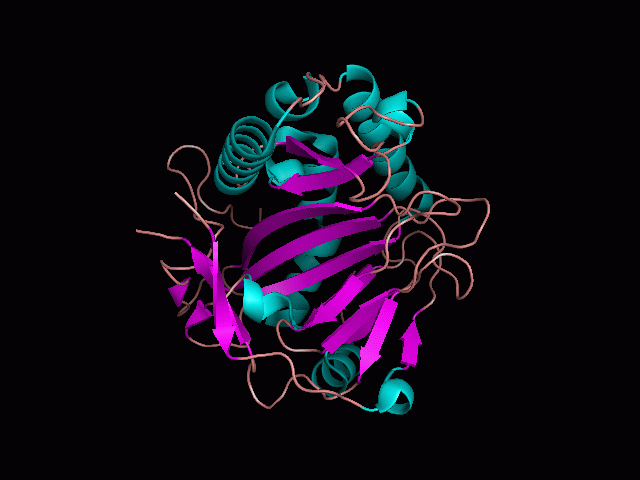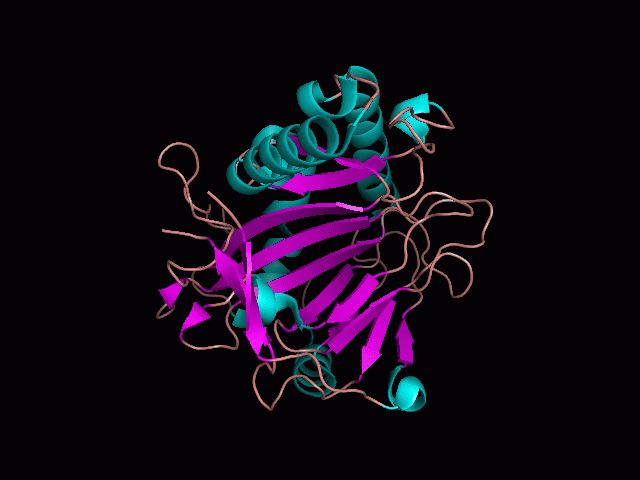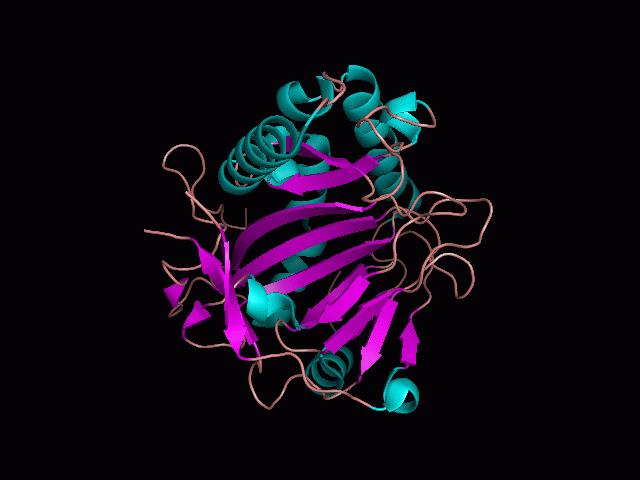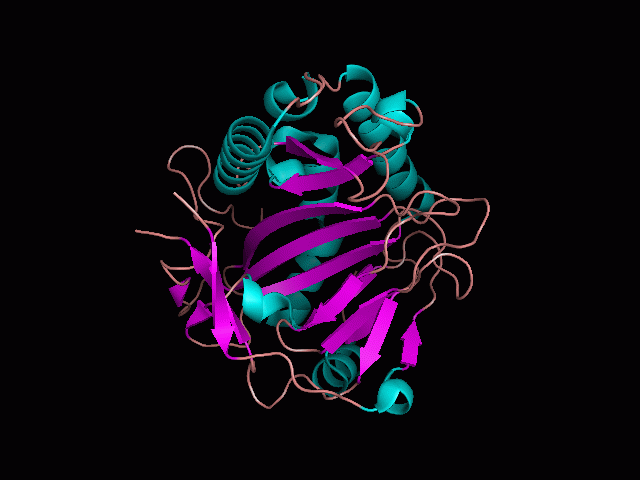
Phytanoyl-CoA dioxygenase Structure
Originally, the structure of our protein was experimentally defined (by Zhang, Z., Butler, D., McDonough, M.A et.al.) via X-ray diffraction.
Name - Phytanoyl-CoA dioxygenase (PHYHD1)
Resolution (Amstrongs) - 1.90
R-Value - 0.221 (obs, relatively low)
Space Group - P 3.1 2 1
Unit Cell Paramters (Amstrongs) - a = 91.97, b = 91.97, c = 81.61
Unit Cell Angles - alpha = 90.00, beta = 90.00, gamma = 120.00
As shown from behind the pocket, there are high levels of steric hinderance if the approaching ligand comes from behind the molecule. This is consisted with functional information, as Liam will show later that the binding sites of the molecule fall within the pocket shown in the following image.
As shown from in front of the pocket, there is a large hole in the structure of our protein. As steric hinderance is low and the surface area is maximized at this position, it would seem that this area should encompass the binding sites of our protein, or at least be the location of some form of interaction. The fact such a definitive hole in the structure of the protein exists lead us to investigate further properties around this region.
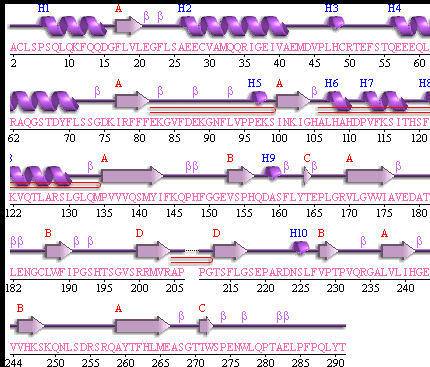
Showing the topology of the protein. Reproduced from the European Bioinformatics Institute,
http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/2opw/domA01.gif
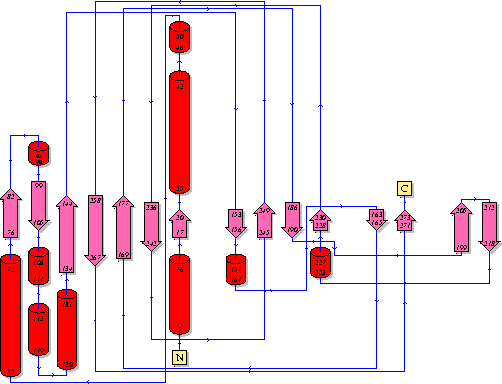
Showing another topology of the protein. Reproduced from the European Bioinformatics Institute,
http://www.ebi.ac.uk/