Pyridoxal Phosphatase Discussion: Difference between revisions
(→PDBsum) |
(→PDBsum) |
||
| Line 47: | Line 47: | ||
* Images of the query structure | * Images of the query structure | ||
* Annotated plots of each protein chain's secondary structure | * Annotated plots of each protein chain's secondary structure | ||
* PROMOTIF-generated structural analyses, | * PROMOTIF-generated structural analyses, illustrated in the form of topology diagrams | ||
* Schematics of protein interactions (i.e. with ligand, DNA), as represented by LIGPLOT | * Schematics of protein interactions (i.e. with ligand, DNA), as represented by LIGPLOT | ||
''(Laskowski et al., 2001)''<BR> | ''(Laskowski et al., 2001)''<BR> | ||
Revision as of 09:47, 9 June 2008
Evolution
It was found that the related proteins had very few conserved regions. One of the regions was _____________. _____________ was also conserved. Several conservative and semi-conservative substitutions were found at __________.
Structure
PDB
Based on the information obtained from PDB, 2cfsA was identified to have the following features:
- Isolated from Homo Sapiens, and is expressed in Escherichia Coli.
- Structurally similar to the Pyridoxal Phosphate Phosphatase protein.
- Consists of a single type of chain (A), and (2) Magnesium components.
- Resolution of 2.4 angstroms. The significance of this is that the probability that the number of side-chains in the wrong rotamer is relatively smaller. Proteins of similar resolution were noted also to: (1) have many small detectable errors, (2) be of correct folding, (3) contain fewer number of errors in the surface loops and (4) consist of visible water molecules and small ligands.
DALI
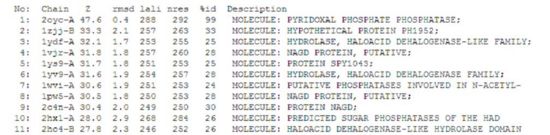
A search on the protein in the Dali database yielded 176 hits, of which the top 11 were identified to be of potential significance on account of the information provided in the summary block of the results. The summary block provides the following information:
- Z score, or the statistical significance of the similarity between the hit protein and the protein-of-interest. The program optimises a weighted sum of similarities of intramolecular distances.
- Root Mean Square Distance (RMSD), which indicates the degree of divergence between the hit protein and the protein-of-interest. The lower the value, the more similar it is to the protein-of-interest.
- lali, the number of structurally equivalent residues.
- nres, or the total number of amino acids in the hit protein.
- %id. As the term implies, %id refers to the percentage of sequence identity over structurally identical positions.
Of the 11 hit proteins, 2oycA was predicted to be the most structurally similar to 2cfsA. This was on virtue of the following properties:
- Among the list of hit proteins generated, it had the highest Z value of 47.6
- RMSD value of 0.4, the lowest among the hit proteins.
- lali value of 288. 2cfsA has a total of 298 amino acid residues, which means that 2oycA and 2cfsA differ by only 10 amino acid residues.
- nres value of 292. This did not bear much significance on the decision-making process. However, a conclusion drawn was that it was similar to 2cfsA in terms of length.
- %id score of 99%, which simply means that based on the information currently stored in the DALI database, 2oycA and 2cfsA were highly similar.
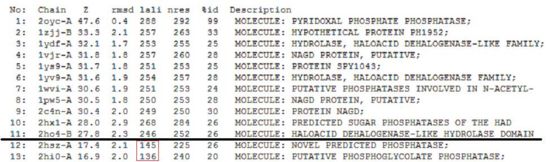
Given that 2cfsA has 298 amino acid residues, the twelfth hit onwards (i.e. 2hszA) were rejected as more than half of their amino acid residues did not indicate similarity to 2cfsA.
To further prove that 2cfsA and 2oycA were structurally similar, their three dimensional structures were superimposed using PyMOL. As expected, the 2oycA bore a close resemblance to 2cfsA, but the differing regions have yet to be identified.
Based on the results obtained in the first (DALI) run, a second run was conducted towards the end of the study. This was primarily due to the concern that the first run identified 2oycA as the query protein instead of 2cfsA. While this would not have (negative) implications on the already-generated results, it was done as a measure of assurance for the team. It was observed that there was a huge deviation between the results obtained in the first and second runs, namely:
- 2cfsA was not among the list of hit proteins yielded in the first run, whereas based on the results obtained in the second run, it was identified as the query protein. This information was significant, as before the second run, it could only be hypothesized that 2cfsA belonged to the same protein family as 2oycA. With the results yielded in the second run, it can be confirmed that 2cfsA and 2oycA were indeed from the same protein family. In fact, the second run yielded a handful of proteins which were more closely related to 2cfsA than 2oycA.
The vast difference between the results obtained in both runs highlighted the disadvantages of DALI. While it is one of the more reliable protein fold comparison programs available, continuous changes in the field has resulted in inconsistencies (Novotny et al., 2004). This has been well illustrated from the results obtained in this study.
Other structural studies indicated the use of the Combinatorial Extension (CE) database. In this study, the CE database was not utilized due to software incompatibility. Whenever a query PDB ID was run against the database, an error message would appear. This could be because CE only accepts structures uploaded from a Mac or a Unix workstation (Novotny et al., 2004), which were not utilized during this study.
PDBsum
PDBsum is a database which pictorially illustrates information on each macromolecule deposited in the PDB. Some of the features provided by PDBsum include:
- Images of the query structure
- Annotated plots of each protein chain's secondary structure
- PROMOTIF-generated structural analyses, illustrated in the form of topology diagrams
- Schematics of protein interactions (i.e. with ligand, DNA), as represented by LIGPLOT
(Laskowski et al., 2001)
The protein chains are represented in the form of "wiring diagrams", a schematic of the protein chain's secondary structure motifs, primary sequence, structural domains and active sites (Laskowski et al., 2001). The secondary structures of 2cfsA and 2oycA were noted to be highly similar, the difference being that 2oycA did not have any disulphide bonds, nor indications of any active site(s), as opposed to 2cfsA.
The topology diagrams of both proteins (2cfsA and 2oycA) were the next bits of information obtained. These topology diagrams are PROMOTIF-generated schematics of the secondary structure motifs, and do not provide information as detailed as that of the "wiring diagrams" described in the previous paragraph (Laskowski et al., 2001).
In this study, the topology diagrams indicated complete homology between the said proteins.
A cleft analysis for both proteins was conducted, and visualization was carried out (via PyMOL) with the aim of observing similarities between the potential catalytic sites of 2cfsA and 2oycA. Clefts in protein surfaces are of relative significance due to their relevance to binding sites. It has been hypothesized that the active site usually lies in the largest protein clefts/cavities (Laskowski et al. 1996).
It was concluded that both proteins may have similar active sites. This is significant as structural information is usually crucial for the functional prediction of the protein-of-interest. Since the information provided by the secondary structures of both proteins are highly similar, there is a great possibility that 2cfsA and 2oycA are functionally similar.
2cfsA was identified to be from the haloacid dehalogenase-like hydrolase family (http://pfam.sanger.ac.uk/family?acc=PF00702). Members of this protein family (which includes phosphatases) belong to the Haloacid Dehalogenase (HAD) superfamily. Almo et al., 2007 mentions that the HAD superfamily encompasses a large number of magnesium-dependent phosphohydrolases characterized by a DXDX motif - which is an indicator of the catalytic site.
PROFUNC
The ProFunc database was utilized due to its ability to predict the likely functions of a protein with a known 3D structure. It makes use of both existing and new methods to analyse a query protein's sequence and structure, with the aim of identifying functional motifs which could indicate relationships to proteins whose functions are known (Laskowski et al., 2005).
Based on the results generated by DALI, it was concluded that 2cfsA and 2oycA were structurally similar and this could have been a crucial point in the functional determination of both proteins. A search on ProFunc, however, seemed to suggest that 2cftA was a much closer match to 2cfsA than 2oycA. In fact, based on the local alignment scoring system, 2cftA is identical to 2cfsA in terms of sequence and structure, according to the information stored in the PDB database. However, this does not rule out the potential significance of 2oycA, as it was the next closest protein hit after 2cftA.
It was important to note that unlike the information provided by NCBI (http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=134104092), 2cfsA was noted to have 293 amino acid residues instead of 298 (NCBI). One reason for this discrepancy could be that in the ever-changing scientific field, existing information will not be spared from change.
In terms of secondary structure matching (SSM), however, 2cftA was NOT identical to 2cfsA, as reflected by their differing Q scores (1.000 and 0.981 respectively). The Q score takes into account the number of aligned residues, the rmsd scores and the size of the proteins; and a high Q score implies high similarity between the hit protein and the protein-of-interest. More interestingly, however, was the observation that 2oycA was a distant sixth on the list, four tiers below 2cftA. Such was the homology between the hit proteins and 2cfsA, however, that even at sixth, the deviation between 2cfsA and 2oycA was minimal.
To determine the potential active sites of 2cfsA, the nest analysis method was utilized. Based on an article by Pal et. al, 2002, the principle of this method revolves around the possibility that anion (negatively-charged) and cation (positively-charged) binding sites in proteins are made up of three amino acids, of which two exhibit "enantiomeric" main chain conformations. This simply means that the main chain torsion angles of the two adjacent amino acids are inverted about the centre of the Ramachandran plot. This results in the formation of "nests", which are defined as concave depressions which ultimately serve as binding sites.
Finally, a LIGPLOT of interactions involving the PLP ligand in 2cftA was obtained. The LIGPLOT was a good indication of the location of 2cftA's active site, highlighting the hydrogen bonds and non-bonded interactions between the ligand (in 2cfsA's case, the Magnesium ions) and the protein residues the ligand interacts with (Laskowski et al., 2001). It was also noticed that the Mg 1296(A) ion of 2cfsA was located in exactly the same position as 2cftA's calcium ion. This is significant as substitution of the Magnesium ion with the catalytically inert Calcium ion results in a loss of activity (Almo et al., 2007). Based on this, it was deduced that it was highly possible for 2cfsA's active site to be in the region surrounding Mg 1296(A). The LIGPLOT of interactions involving the Mg 1297(A) ion of 2cfsA was obtained, and this information - along with the information provided by the LIGPLOT involving the Mg 1296(A) - was used to generated a three-dimensional view of the catalytic site of 2cfsA via PyMOL. Compared to the earlier results based on the evolutionary aspect of the paper, the conserved regions - as obtained from the Multiple Sequence Alignment, was not too far off from the information (residues that constitute the catalytic site) obtained from Almo et al., 2007. PyMOL-generated visualizations of the possible catalytic sites using both sets of information illustrated this.