Result of SNAPG: Difference between revisions
No edit summary |
|||
| Line 1: | Line 1: | ||
== SNAPG Structure == | == SNAPG Structure == | ||
=== Structure architecture === | === Structure architecture === | ||
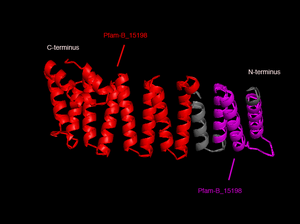
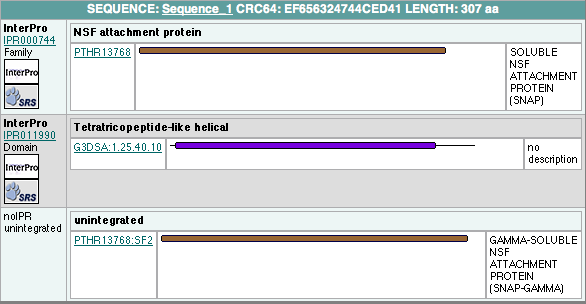
In order to analyze protein structure of SNAPG, structural comparison to known protein structure is required. An insight to SNAPG structural arrangement provides various informative data on possible protein functions and interactions with another protein and/or DNA. Based on protein families database, Pfam at Sanger, it was found that SNAPG protein matched to Pfam-B protein families and consist of 2 domains, Pfam-B_7270 (PB007270) and Pfam-B_15198 (PB015189) respectively as shown in Figure 1. Both of 2 domains appears to be associated with NSF attachment protein activity [http://www.ebi.ac.uk/interpro/IEntry?ac=IPR000744]. | In order to analyze protein structure of SNAPG, structural comparison to known protein structure is required. An insight to SNAPG structural arrangement provides various informative data on possible protein functions and interactions with another protein and/or DNA. Based on protein families database, Pfam at Sanger, it was found that SNAPG protein matched to Pfam-B protein families and consist of 2 domains, Pfam-B_7270 (PB007270) and Pfam-B_15198 (PB015189) respectively as shown in Figure 1. Both of 2 domains appears to be associated with NSF attachment protein activity [http://www.ebi.ac.uk/interpro/IEntry?ac=IPR000744]. | ||
Revision as of 01:12, 11 June 2007
SNAPG Structure
Structure architecture
In order to analyze protein structure of SNAPG, structural comparison to known protein structure is required. An insight to SNAPG structural arrangement provides various informative data on possible protein functions and interactions with another protein and/or DNA. Based on protein families database, Pfam at Sanger, it was found that SNAPG protein matched to Pfam-B protein families and consist of 2 domains, Pfam-B_7270 (PB007270) and Pfam-B_15198 (PB015189) respectively as shown in Figure 1. Both of 2 domains appears to be associated with NSF attachment protein activity [1].
Structural comparison
Dali webserver is one of the powerful tool to screen any protein that are structurally homologous with our query. Two structurally related proteins with highest Z-value generated by Dali server were chosen for SNAPG structure comparison analysis. These proteins were vesicular transport ptotein sec17 (1qqe) and type 4 fimbrial biogenesis protein (2f17) (refer to table..)
| PDB-chain | Structure | Z-value | % identity | Protein |
|---|---|---|---|---|
| 2IFU-A | 37.8 | 100 | Endocytosis/exocytosis. Gamma-SNAP (Danio rerio) | |
| 1QQE-A | 23.3 | 23 | Protein binding. Vesicular transport protein sec17(yeast) | |
| 2FI7-A | 12.9 | 14 | Protein transport. Type 4 fimbrial biogenesis protein pili (Pseudomonas aeruginosa) |