Structural analysis of 2qgn: Difference between revisions
Choojinhsien (talk | contribs) |
Choojinhsien (talk | contribs) |
||
| Line 45: | Line 45: | ||
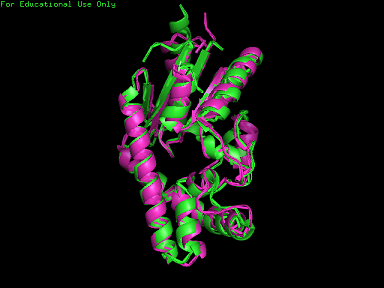
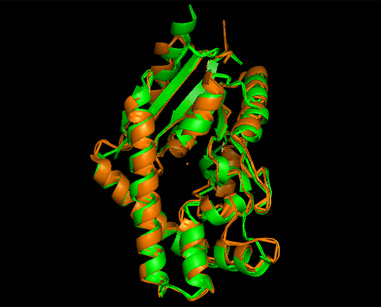
A total of 527 hits were found from DALI search, nonetheless only the first 20 hits that may be of significance were shown on the figure. Based on the outcome of DALI, PDB files of each structurally similar protein was obtained from PDB. These were each superimposed against 2qgn using the PyMOL software, to compare the structural similiarity. Results are as below : | A total of 527 hits were found from DALI search, nonetheless only the first 20 hits that may be of significance were shown on the figure. Based on the outcome of DALI, PDB files of each structurally similar protein was obtained from PDB. These were each superimposed against 2qgn using the PyMOL software, to compare the structural similiarity. Results are as below : | ||
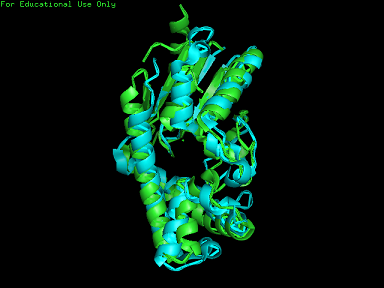
[[Image:2qgn.png|framed|none]][[Image:2qgn2.png|framed|none]][[Image:2qgn421.jpg|framed|none]] | [[Image:2qgn.png|framed|'''Figure 7'''2ze5 superimposed against 2qgn via PyMOL.|none]] | ||
[[Image:2qgn2.png|framed|'''Figure 8'''3crm superimposed against 2qgn via PyMOL.|none]] | |||
[[Image:2qgn421.jpg|framed|'''Figure 9'''3crq superimposed against 2qgn via PyMOL.|none]] | |||
Revision as of 14:21, 31 May 2008
Protein Sequence in FASTA format
>gi|152149497|pdb|2QGN|A Chain A, Crystal Structure Of Trna Isopentenylpyrophosphate Transferase (Bh2366) From Bacillus Halodurans, Northeast Structural Genomics Consortium Target Bhr41. XKEKLVAIVGPTAVGKTKTSVXLAKRLNGEVISGDSXQVYRGXDIGTAKITAEEXDGVPHHLIDIKDPSE SFSVADFQDLATPLITEIHERGRLPFLVGGTGLYVNAVIHQFNLGDIRADEDYRHELEAFVNSYGVQALH DKLSKIDPKAAAAIHPNNYRRVIRALEIIKLTGKTVTEQARHEEETPSPYNLVXIGLTXERDVLYDRINR RVDQXVEEGLIDEAKKLYDRGIRDCQSVQAIGYKEXYDYLDGNVTLEEAIDTLKRNSRRYAKRQLTWFRN KANVTWFDXTDVDFDKKIXEIHNFIAGKLEEKSKLEHHHHHH
Structure of Protein
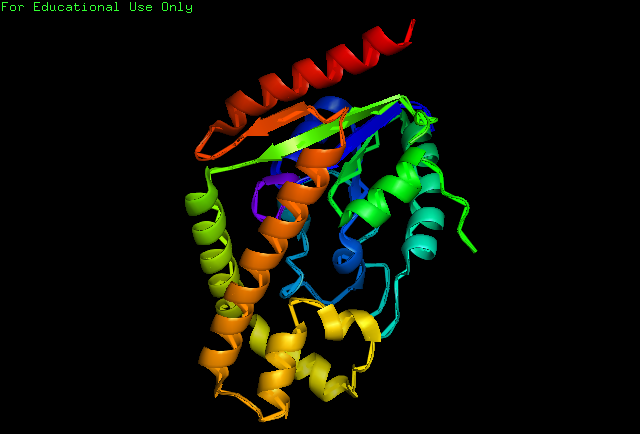
Secondary Structure
Analysis of the secondary structure acquired from Protein Data Bank showed results as displayed below :
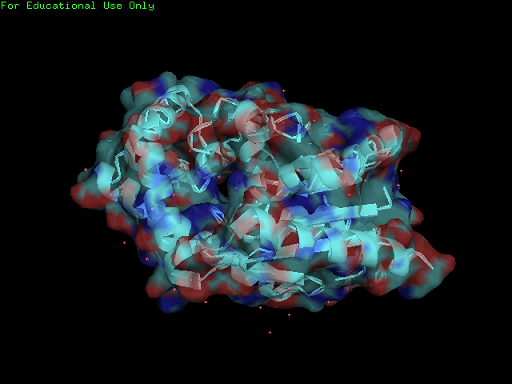
Surface Properties
Domains
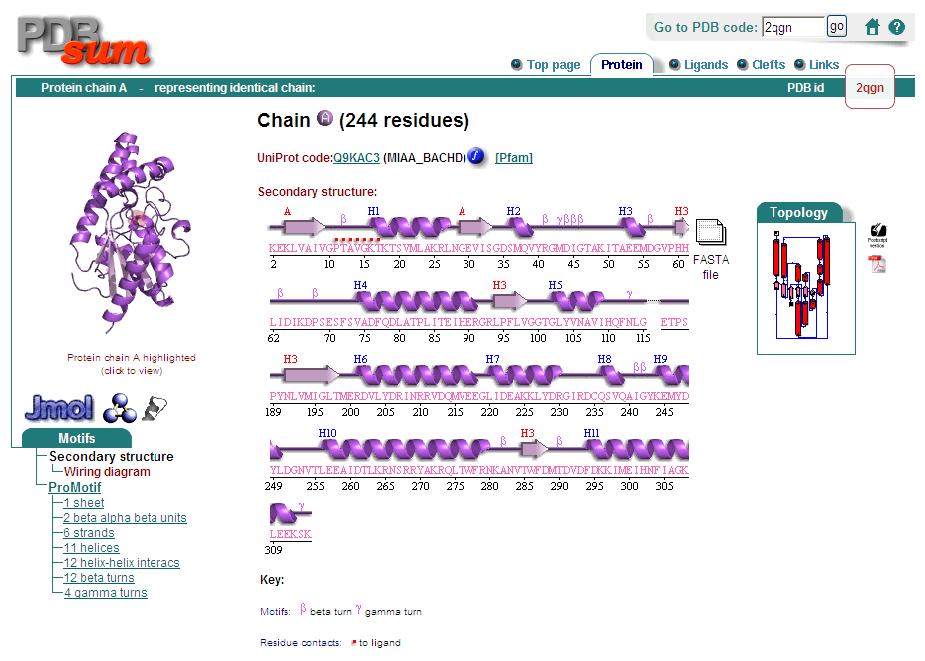
2qgnA is composed of two main domains. CATH analysis of 2qgn resulted in the finding of two main domains composing 2qgnA.
One domains ranges from residue 2-200 and residue 283-314. Another domain encompasses residues from 201-282.
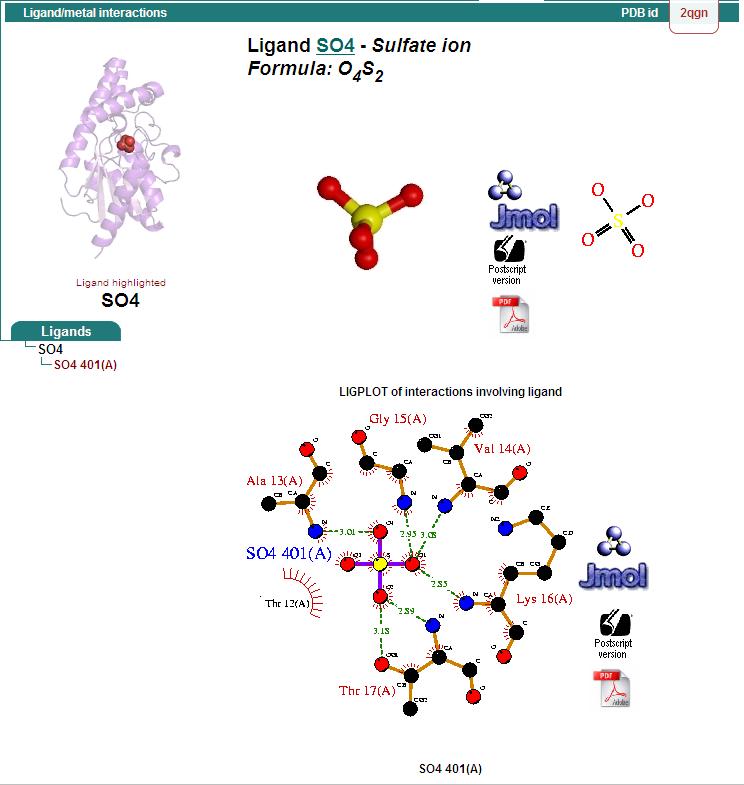
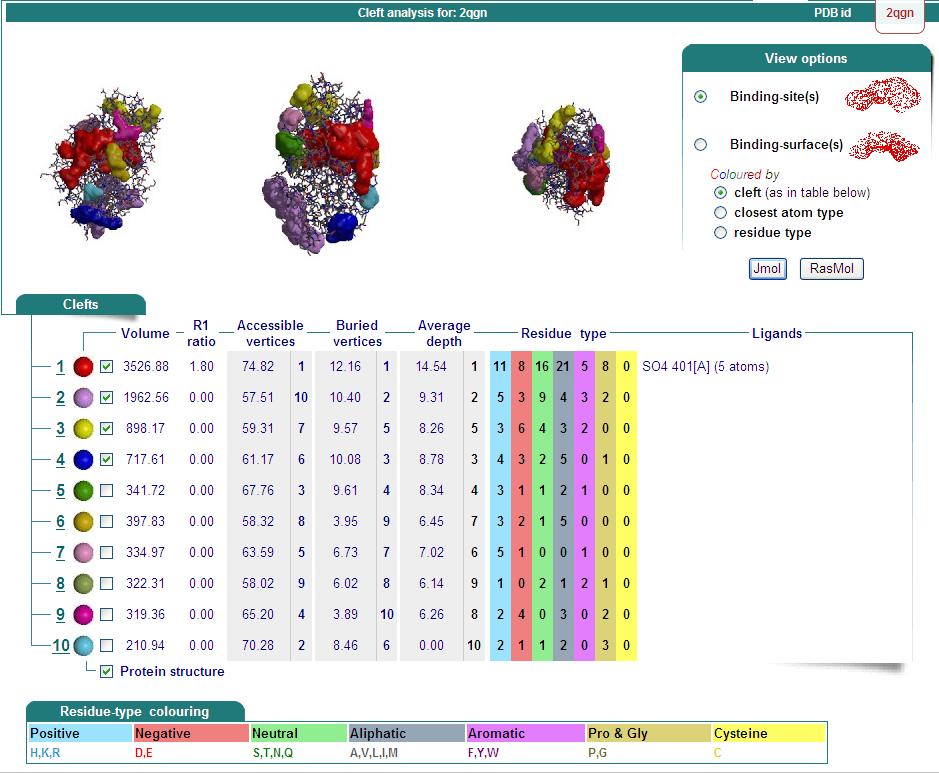
Ligand Binding Sites and Surface Clefts
Structural Alignment
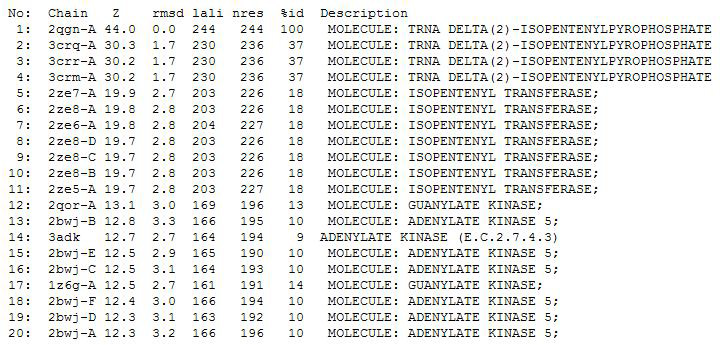
Dali Output
PDB entry code for 2qgn was loaded onto DALI to search for structurally similar neighbours. Displayed below are the results from DALI search :-
A total of 527 hits were found from DALI search, nonetheless only the first 20 hits that may be of significance were shown on the figure. Based on the outcome of DALI, PDB files of each structurally similar protein was obtained from PDB. These were each superimposed against 2qgn using the PyMOL software, to compare the structural similiarity. Results are as below :