Structure: Kenn: Difference between revisions
From MDWiki
Jump to navigationJump to search
No edit summary |
No edit summary |
||
| Line 11: | Line 11: | ||
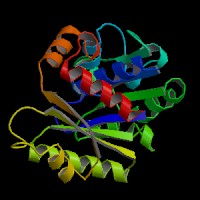
[[Image:NUBP2BACK.jpg|left|frame|NUBP2 Front view]][[Image:nubp2front.jpg| | [[Image:NUBP2BACK.jpg|left|frame|NUBP2 Front view]][[Image:nubp2front.jpg|right|frame|NUBP2 Top view]][[Image:nubp2top.jpg|none|frame|NUBP2 Back view]] | ||
| Line 17: | Line 17: | ||
---- | ---- | ||
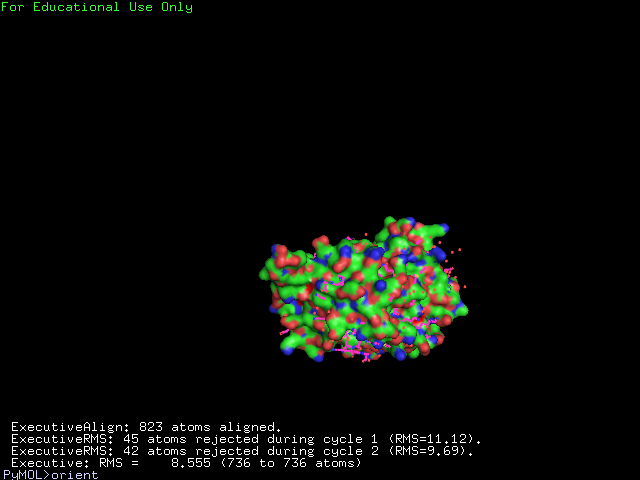
[[Image:alignment1g3p.jpg|400px|frame|The alignment of 2ph1 and 1g3q]] | [[Image:alignment1g3p.jpg|400px|center|frame|The alignment of 2ph1 and 1g3q]] | ||
The structure of 1g3q is the most similar to 2ph1 according to the result from Dali.This figure shows the alignment of 2ph1 and 1g3q with RMS 0.55. | The structure of 1g3q is the most similar to 2ph1 according to the result from Dali.This figure shows the alignment of 2ph1 and 1g3q with RMS 0.55. | ||
---- | ---- | ||
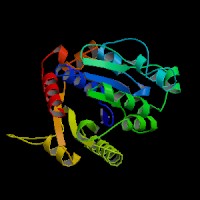
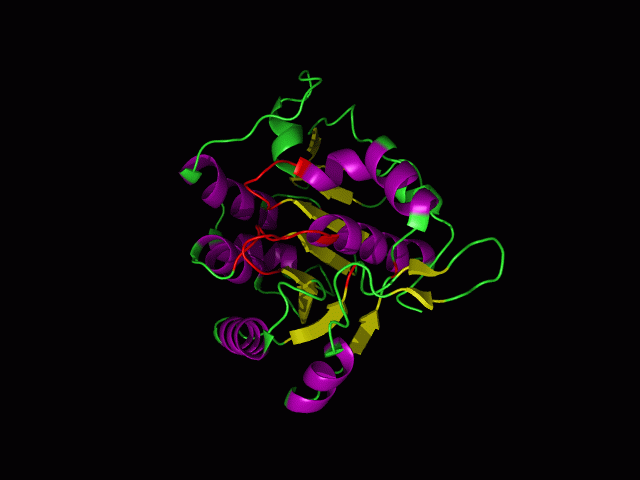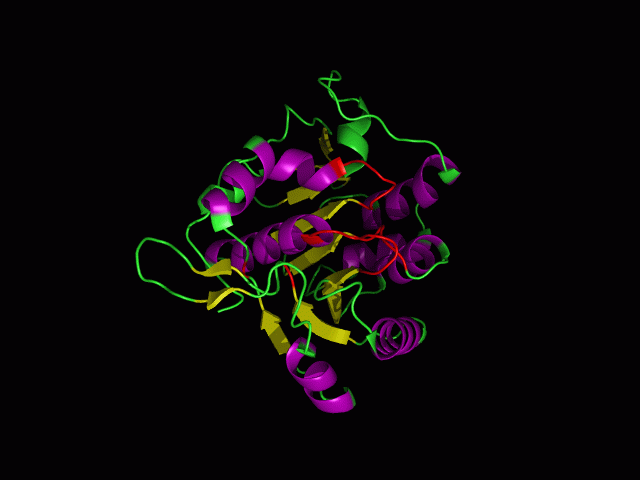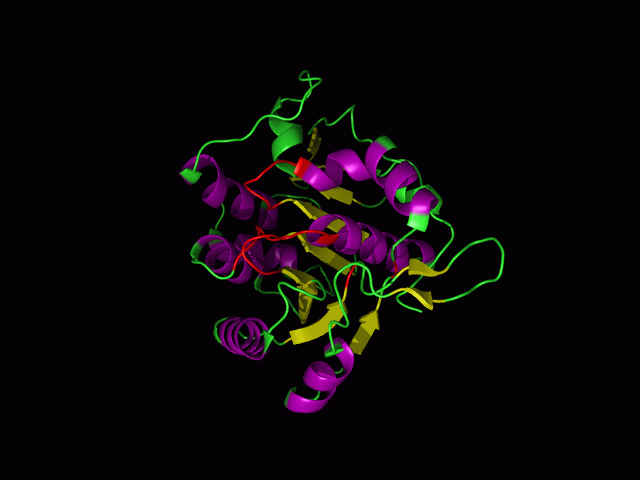
[[Image:movie.jpg|frame|The secondary structure of 2ph1]] | [[Image:movie.jpg|center|frame|The secondary structure of 2ph1]] | ||
This picture shows the secondary structure of 2ph1, the alpha helixes were shown in purple, and the beta sheets are shown in yellow. | This picture shows the secondary structure of 2ph1, the alpha helixes were shown in purple, and the beta sheets are shown in yellow. | ||
---- | ---- | ||
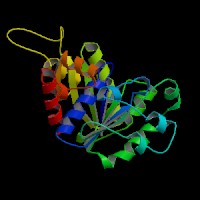
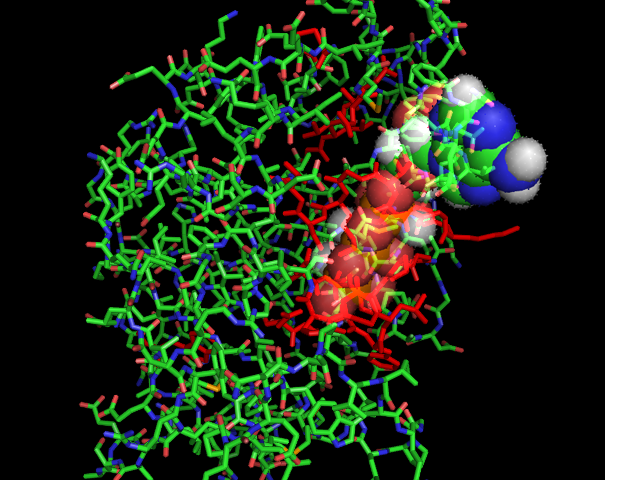
[[Image:ATPbinding.jpg|frame|ATP binding to 2ph1]] | [[Image:ATPbinding.jpg|center|frame|ATP binding to 2ph1]] | ||
The ATP molocule was binding to the p-loop of 2ph1. | The ATP molocule was binding to the p-loop of 2ph1. | ||
Revision as of 16:04, 7 June 2008
The P-loop connecting b1 and a1, has well conserved sequences, and its amino acid side chains exhibit the same conformations among all other proteins with the GXXXXGKT motif.Other conserved motifs are also observed in MinD. The Walker B motif (residues 118±121) is located near the terminus of b5, and has the modi®ed sequence, Asp-XPro- Ala.
The structure of 1g3q is the most similar to 2ph1 according to the result from Dali.This figure shows the alignment of 2ph1 and 1g3q with RMS 0.55.
This picture shows the secondary structure of 2ph1, the alpha helixes were shown in purple, and the beta sheets are shown in yellow.
The ATP molocule was binding to the p-loop of 2ph1.