Structure: Kenn: Difference between revisions
No edit summary |
No edit summary |
||
| (11 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== '''Structure Summary'''== | |||
NESG ID: GR165 | |||
PDB ID: 2ph1 | |||
Deposition date: 2007-04-10 | |||
Common Name: Nucleotide-binding protein | |||
Length (a.a.): 240 | |||
Organism: Archaeoglobus fulgidus | |||
SwissProt / TrEMBL ID: O30288_ARCFU | |||
Oligomerization: monomer | |||
Molecular weight: 26087 | |||
Secondary Structure Elements: | |||
alpha helices: 7-14, 32-45, 61-65, 111-122, 143-151, 167-178, 211-217, 230-237 | |||
beta strands: 83-84, 91-93, 50-54, 131-135, 20-24, 155-160, 184-189, 222-225, 74-76, 79-81, 194-195, 202-203 | |||
Resolution: 2.70 Å R-factor: 0.217 R-free: 0.278 | |||
---- | |||
'''DaliLite: Structural Neighbours''' | |||
'''Query: mol1A MOLECULE: NUCLEOTIDE-BINDING PROTEIN; ''' | |||
Matches are sorted by Z-score. Similarities with a Z-score lower than 2 are spurious. | |||
'''Summary''' | |||
No: Chain Z rmsd lali nres %id Description | |||
1: 2ph1-A 47.3 0.0 247 247 100 MOLECULE: NUCLEOTIDE-BINDING PROTEIN; | |||
2: 1g3q-A 20.0 2.7 207 237 21 MOLECULE: CELL DIVISION INHIBITOR; | |||
3: 1ion-A 19.8 2.6 206 243 20 MOLECULE: PROBABLE CELL DIVISION INHIBITOR MIND; | |||
4: 1hyq-A 19.6 3.1 197 232 25 MOLECULE: CELL DIVISION INHIBITOR (MIND-1); | |||
5: 1g3r-A 19.6 2.7 206 237 20 MOLECULE: CELL DIVISION INHIBITOR; | |||
6: 1cp2-A 19.0 2.8 204 269 16 MOLECULE: NITROGENASE IRON PROTEIN; | |||
7: 1g1m-B 18.9 2.8 202 289 18 MOLECULE: NITROGENASE IRON PROTEIN; | |||
8: 1xd9-A 18.8 2.8 203 289 17 MOLECULE: NITROGENASE IRON PROTEIN 1; | |||
9: 1cp2-B 18.8 2.7 203 269 16 MOLECULE: NITROGENASE IRON PROTEIN; | |||
10: 1nip-B 18.4 2.8 202 287 18 NITROGENASE IRON PROTEIN | |||
11: 1de0-A 18.4 2.8 203 289 18 MOLECULE: NITROGENASE IRON PROTEIN; | |||
12: 1xdb-A 18.3 2.9 204 289 18 MOLECULE: NITROGENASE IRON PROTEIN 1; | |||
13: 1de0-B 18.3 2.8 203 289 18 MOLECULE: NITROGENASE IRON PROTEIN; | |||
14: 2bej-A 18.1 3.0 202 245 26 MOLECULE: SEGREGATION PROTEIN; | |||
15: 1n2c-F 18.1 2.9 203 274 18 MOLECULE: NITROGENASE MOLYBDENUM-IRON PROTEIN; | |||
16: 1n2c-E 18.1 3.0 205 274 18 MOLECULE: NITROGENASE MOLYBDENUM-IRON PROTEIN; | |||
17: 1g5p-B 18.1 2.7 202 289 18 MOLECULE: NITROGENASE IRON PROTEIN; | |||
18: 1g5p-A 18.1 2.8 201 286 17 MOLECULE: NITROGENASE IRON PROTEIN; | |||
19: 1g1m-A 18.1 2.8 202 287 18 MOLECULE: NITROGENASE IRON PROTEIN; | |||
20: 2nip-B 18.0 2.7 201 289 18 MOLECULE: NITROGENASE IRON PROTEIN; | |||
21: 2nip-A 18.0 2.8 201 286 17 MOLECULE: NITROGENASE IRON PROTEIN; | |||
22: 1nip-A 18.0 2.8 203 283 18 NITROGENASE IRON PROTEIN | |||
23: 1n2c-G 18.0 2.9 203 274 18 MOLECULE: NITROGENASE MOLYBDENUM-IRON PROTEIN; | |||
24: 1m34-F 18.0 2.8 201 274 18 MOLECULE: NITROGENASE MOLYBDENUM-IRON PROTEIN ALPHA CHAIN; | |||
---- | ---- | ||
[[Image: | |||
''EMI result'' | |||
[[Image:EMIi.jpg|center]] | |||
---- | |||
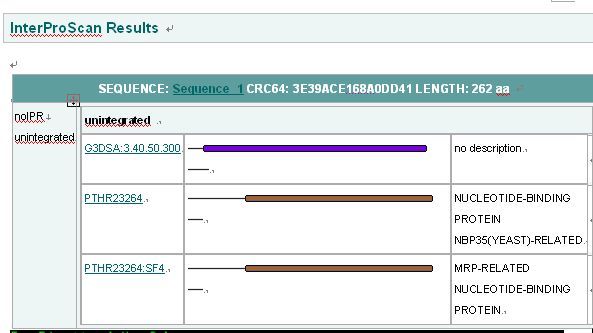
''Interpro Result'' | |||
[[Image:INTERPRO.jpg|center]] | |||
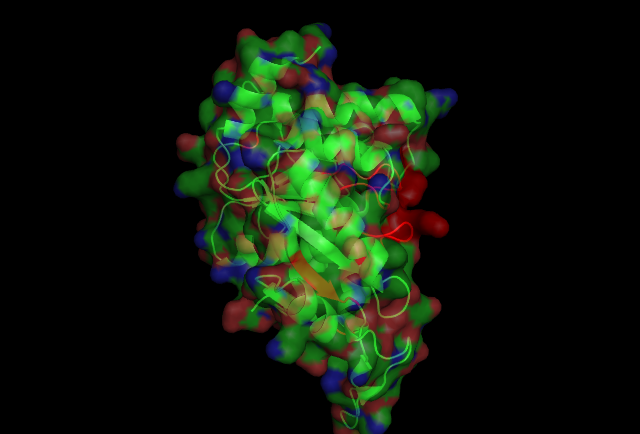
[[Image:2ph1 surface.jpg|400px|frame|The P-loop connecting | |||
b1 and a1, has well conserved sequences, and its amino | |||
acid side chains exhibit the same conformations among all | |||
other proteins with the GXXXXGKT motif.Other conserved motifs are also observed in MinD. The | |||
Walker B motif (residues 118±121) is located near the | |||
terminus of b5, and has the modi®ed sequence, Asp-XPro- | |||
Ala.]] | |||
---- | ---- | ||
[[Image:alignment1g3p.jpg|400px]] | |||
[[Image:NUBP2BACK.jpg|left|frame|NUBP2 Front view]][[Image:nubp2front.jpg|right|frame|NUBP2 Top view]][[Image:nubp2top.jpg|none|frame|NUBP2 Back view]] | |||
---- | |||
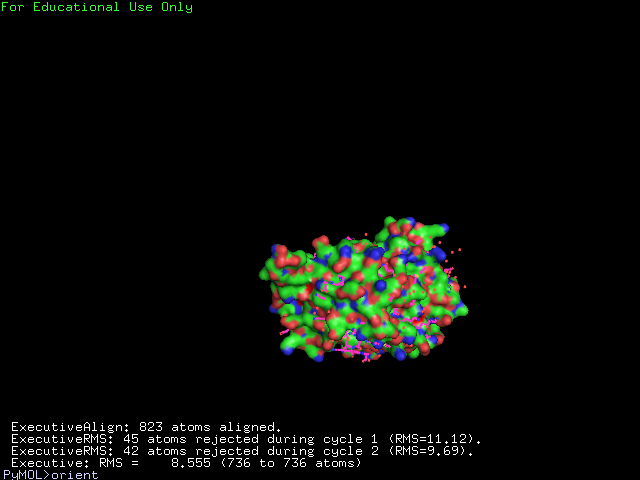
[[Image:alignment1g3p.jpg|400px|center|frame|The alignment of 2ph1 and 1g3q]] | |||
The structure of 1g3q is the most similar to 2ph1 according to the result from Dali.This figure shows the alignment of 2ph1 and 1g3q with RMS 0.55. | |||
---- | |||
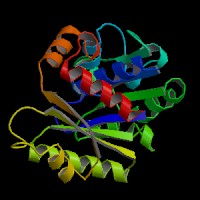
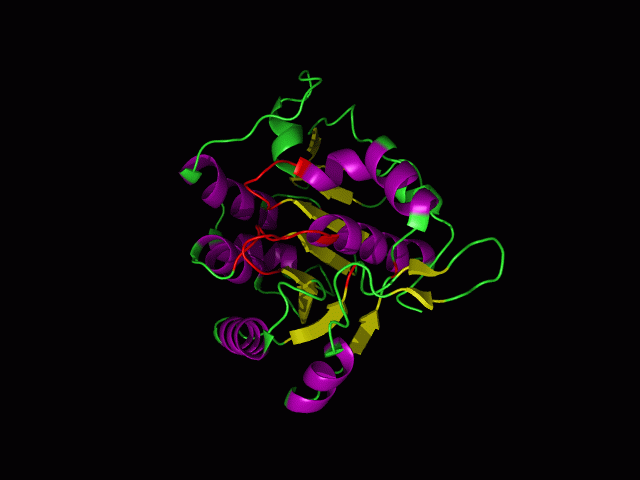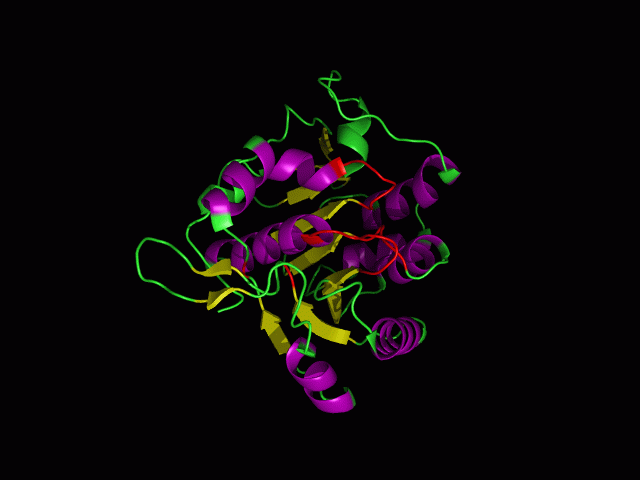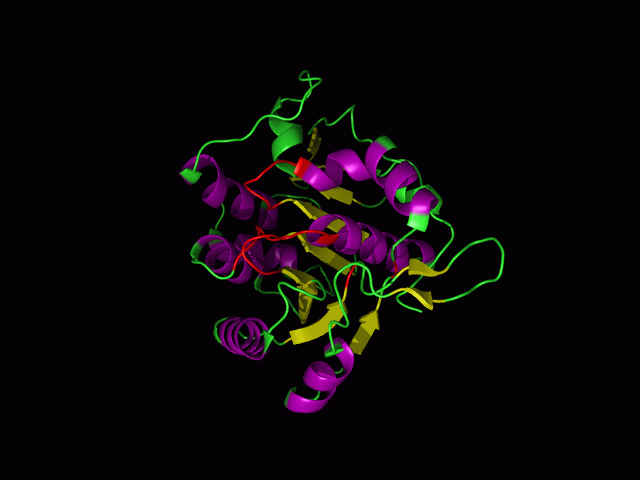
[[Image:movie.jpg|center|frame|The secondary structure of 2ph1]] | |||
This picture shows the secondary structure of 2ph1, the alpha helixes were shown in purple, and the beta sheets are shown in yellow. | |||
---- | |||
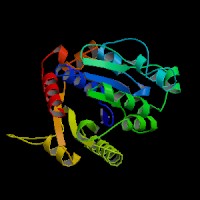
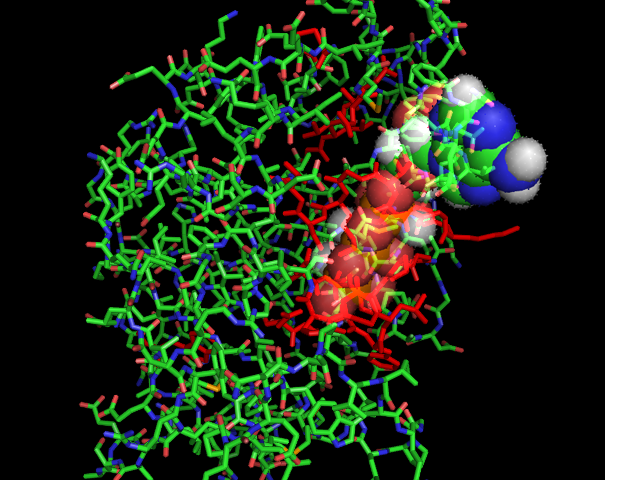
[[Image:ATPbinding.jpg|center|frame|ATP binding to 2ph1]] | |||
The ATP molocule was binding to the p-loop of 2ph1. | |||
---- | |||
---- | ---- | ||
Latest revision as of 17:28, 7 June 2008
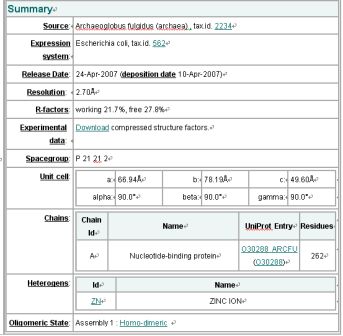
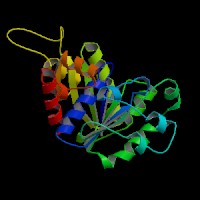
Structure Summary
NESG ID: GR165 PDB ID: 2ph1 Deposition date: 2007-04-10 Common Name: Nucleotide-binding protein Length (a.a.): 240 Organism: Archaeoglobus fulgidus SwissProt / TrEMBL ID: O30288_ARCFU Oligomerization: monomer Molecular weight: 26087
Secondary Structure Elements: alpha helices: 7-14, 32-45, 61-65, 111-122, 143-151, 167-178, 211-217, 230-237 beta strands: 83-84, 91-93, 50-54, 131-135, 20-24, 155-160, 184-189, 222-225, 74-76, 79-81, 194-195, 202-203
Resolution: 2.70 Å R-factor: 0.217 R-free: 0.278
DaliLite: Structural Neighbours
Query: mol1A MOLECULE: NUCLEOTIDE-BINDING PROTEIN;
Matches are sorted by Z-score. Similarities with a Z-score lower than 2 are spurious.
Summary
No: Chain Z rmsd lali nres %id Description
1: 2ph1-A 47.3 0.0 247 247 100 MOLECULE: NUCLEOTIDE-BINDING PROTEIN;
2: 1g3q-A 20.0 2.7 207 237 21 MOLECULE: CELL DIVISION INHIBITOR;
3: 1ion-A 19.8 2.6 206 243 20 MOLECULE: PROBABLE CELL DIVISION INHIBITOR MIND;
4: 1hyq-A 19.6 3.1 197 232 25 MOLECULE: CELL DIVISION INHIBITOR (MIND-1);
5: 1g3r-A 19.6 2.7 206 237 20 MOLECULE: CELL DIVISION INHIBITOR;
6: 1cp2-A 19.0 2.8 204 269 16 MOLECULE: NITROGENASE IRON PROTEIN;
7: 1g1m-B 18.9 2.8 202 289 18 MOLECULE: NITROGENASE IRON PROTEIN;
8: 1xd9-A 18.8 2.8 203 289 17 MOLECULE: NITROGENASE IRON PROTEIN 1;
9: 1cp2-B 18.8 2.7 203 269 16 MOLECULE: NITROGENASE IRON PROTEIN;
10: 1nip-B 18.4 2.8 202 287 18 NITROGENASE IRON PROTEIN
11: 1de0-A 18.4 2.8 203 289 18 MOLECULE: NITROGENASE IRON PROTEIN;
12: 1xdb-A 18.3 2.9 204 289 18 MOLECULE: NITROGENASE IRON PROTEIN 1;
13: 1de0-B 18.3 2.8 203 289 18 MOLECULE: NITROGENASE IRON PROTEIN;
14: 2bej-A 18.1 3.0 202 245 26 MOLECULE: SEGREGATION PROTEIN;
15: 1n2c-F 18.1 2.9 203 274 18 MOLECULE: NITROGENASE MOLYBDENUM-IRON PROTEIN;
16: 1n2c-E 18.1 3.0 205 274 18 MOLECULE: NITROGENASE MOLYBDENUM-IRON PROTEIN;
17: 1g5p-B 18.1 2.7 202 289 18 MOLECULE: NITROGENASE IRON PROTEIN;
18: 1g5p-A 18.1 2.8 201 286 17 MOLECULE: NITROGENASE IRON PROTEIN;
19: 1g1m-A 18.1 2.8 202 287 18 MOLECULE: NITROGENASE IRON PROTEIN;
20: 2nip-B 18.0 2.7 201 289 18 MOLECULE: NITROGENASE IRON PROTEIN;
21: 2nip-A 18.0 2.8 201 286 17 MOLECULE: NITROGENASE IRON PROTEIN;
22: 1nip-A 18.0 2.8 203 283 18 NITROGENASE IRON PROTEIN
23: 1n2c-G 18.0 2.9 203 274 18 MOLECULE: NITROGENASE MOLYBDENUM-IRON PROTEIN;
24: 1m34-F 18.0 2.8 201 274 18 MOLECULE: NITROGENASE MOLYBDENUM-IRON PROTEIN ALPHA CHAIN;
EMI result
Interpro Result
The structure of 1g3q is the most similar to 2ph1 according to the result from Dali.This figure shows the alignment of 2ph1 and 1g3q with RMS 0.55.
This picture shows the secondary structure of 2ph1, the alpha helixes were shown in purple, and the beta sheets are shown in yellow.
The ATP molocule was binding to the p-loop of 2ph1.