Structure of 1zkd: Difference between revisions
No edit summary |
No edit summary |
||
| Line 151: | Line 151: | ||
Located on 17E3 on mouse chromosome with 436 aa | Located on 17E3 on mouse chromosome with 436 aa | ||
[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=11746687 Article on 1im8] | [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=11746687 Article on 1im8] | ||
Most probably a methyltransferase as most of the matches from DALI are transferase | Most probably a methyltransferase as most of the matches from DALI are transferase | ||
[[Image:InterPro.JPG|InterPro Scan Results|Left]] | [[Image:InterPro.JPG|InterPro Scan Results|Left]] | ||
Revision as of 14:32, 4 June 2007
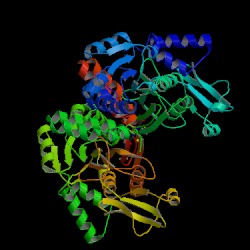
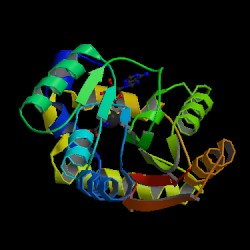
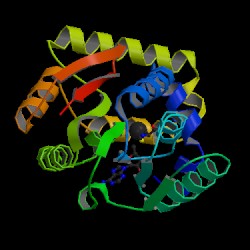
Result from DALI shows proteins that are similar to the 1zkd. They are 2ex4 and 1im8. These 2 proteins matches 1zkd the most according to DALI. The pictures below shows the structures of the proteins related comparing to 1zkd.
With these informations we can now look into the proteins that is similar to 1zkd and predict the functions. 1im8 is found to be a methyltransferase with a bound S-adenosylhomocysteine from the crystal structure of YecO from Haemophilus influenzae (HI0319). 2ex4is a human methyltransferase with S-adenosylhomocysteine.
Alignment with 2ex4 1ZKD:A 24/25 WRYXELCLGHPEHGYYV--TRDPLGREGDFTTSPEISQXFGELLGLWSASVWKAAD-EPQ 2EX4:A 24/7 IEDEKQFYS----KAKTYWKQIPPTVDGMLGGYGHISSIDINSSRKFLQRFLREGPNKTG 1ZKD:A 81/82 TLRLIEIGPGRGTXXADALRALRVLPILYQSLSVHLVEINPVLRQKQQTLLAGI-RNIHW 2EX4:A 80/64 TSCALDCGAGIGRITKRLLLPL--------FREVDMVDITEDFLVQAKTYLGEEGKRVRN 1ZKD:A 140/141 HD-----SFEDVPEGPAVILANEYFDVLPIHQAIKRETGWHERVIEIGASGELVFGVAAD 2EX4:A 132/116 YFCCGLQDFTPEPDSYDVIWIQWVIGHLT------------------------------- 1ZKD:A 195/196 PIPGFEALLPPLARLSPPGAVFEWRP--DTEILKIASRVRDQGGAALIIDYG--HLRSDV 2EX4:A 161/145 ------------------------DQHLAEFLRRCKGSL-RPNGIIVIKDNMAQE----- 1ZKD:A 251/252 GDTFQAIASHSYADPLQHPGRADLTAHV---DFDALGRAAESIGARAHGPVTQG 2EX4:A 191/175 GVILDD---------------VDSSVCRDLDVVRRIICSAG---LSLLAEERQE
Alignment with 1im8 1ZKD:A 59/60 QXFGELLGLWSASVWKAADEPQTLRLIEIGPGRGTXXADALRALRVLPILYQSLSVHLVE 1IM8:A 42/40 SNIITAIGXLAERFV-----TADSNVYDLGCSRGAATLSARRNI-----NQPNVKIIGID 1ZKD:A 119/120 INPVLRQKQQTLLAGI---RNIHWHD--SFEDVPEGPAVILANEYFDVLPIHQAIKRETG 1IM8:A 92/90 NSQPXVERCRQHIAAYHSEIPVEILCNDIRHVEIKNASXVILNFTLQFLP---------- 1ZKD:A 174/175 WHERVIEIGASGELVFGVAADPIPGFEALLPPLARLSPPGAVFEWRP--DTEILKIASRV 1IM8:A 142/140 ---------------------------------------------PEDRIALLTKIYEGL 1ZKD:A 232/233 RDQG--GAALIIDYG 1IM8:A 157/155 ---NPNGVLVLSEKF
Ligang Binding Sites
S-ADENOSYL-L-HOMOCYSTEINEis found on the 2 methyltranferases.
Sequence
MIDQTALATEIKRLIKAAGPMPVWRYMELCLGHPEHGYYVTRDPLGREGDFTTSPEISQMFGELLGLWSASVWKAADEPQ TLRLIEIGPGRGTMMADALRALRVLPILYQSLSVHLVEINPVLRQKQQTLLAGIRNIHWHDSFEDVPEGPAVILANEYFD VLPIHQAIKRETGWHERVIEIGASGELVFGVAADPIPGFEALLPPLARLSPPGAVFEWRPDTEILKIASRVRDQGGAALI IDYGHLRSDVGDTFQAIASHSYADPLQHPGRADLTAHVDFDALGRAAESIGARAHGPVTQGAFLKRLGIETRALSLMAKA TPQVSEDIAGALQRLTGEGRGAMGSMFKVIGVSDPKIETLVALSDDTDREAERRQGTHGLEHHHHHH
Information on 1zkd
Located on 2p22.2 on human chromosome with 441 aa
Located on 17E3 on mouse chromosome with 436 aa
Most probably a methyltransferase as most of the matches from DALI are transferase
Pfam Results
DUF185: domain 1 of 1, from 64 to 299: score 227.1, E = 3.9e-65
*->alArwllveykllgyPYadlnlvElGaGrGtaielmsdlLryiarlv
+l++w + ++k+ ++P l+l E+G+GrGt +m+d+Lr+ r+
query 64 LLGLWSASVWKAADEP-QTLRLIEIGPGRGT---MMADALRA-LRVL 105
PdvyartryylvEiSprLaarQketLapkvaplGhdskveieatdlsglv
P +y+ ++++lvEi+p L+++Q++ La ++ ++
query 106 PILYQSLSVHLVEINPVLRQKQQTLLA-----------------GIR-NI 137
rWhdasileedPdgvptvliaNEVlDalPHDlvrfdkrgggwyErhVlvd
Whd s +e++P+g p v++aNE +D lP +++ +kr+ gw+Er V ++
query 138 HWHD-S-FEDVPEG-PAVILANEYFDVLP--IHQAIKRETGWHER-V-IE 180
ldgdfrlvysqeldplaglaltlreaaldPVKstkklvpsalskllpkll
+ ++lv+++++dp g+ ++l
query 181 IGASGELVFGVAADPIPGFEAL------------------------LPPL 206
ppaeevgygtEvYsParllellqalaerLpahrGrlLaiDYGhlaseyyh
+ +g+++E+ P e+l+++ + + +G++L+iDYGhl+s
query 207 ARLSPPGAVFEW-RPDT--EILKIASRVRD-QGGAALIIDYGHLRSD--- 249
prrksalaaemfngtllqayrqhahddpltnpssllVlyStvaqGlaDiT
g+++qa+ h + dpl +p G+aD+T
query 250 ------------VGDTFQAIASHSYADPLQHP------------GRADLT 275
ahVDFtalaradqyqtaakaagdlkvlgvet<-*
ahVDF+al +aa +g + + g+ t
query 276 AHVDFDALG------RAAESIG-ARAHGPVT 299