Structure of 1zkd: Difference between revisions
No edit summary |
No edit summary |
||
| Line 10: | Line 10: | ||
[[Image:1im8 bio r 251.jpg|thumb|1im8|left]] | [[Image:1im8 bio r 251.jpg|thumb|1im8|left]] | ||
Revision as of 05:49, 29 May 2007
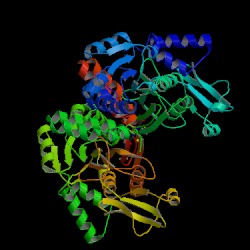
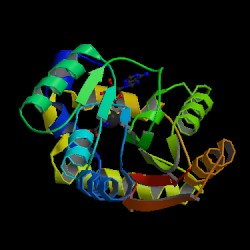
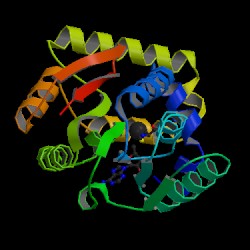
Result from DALI shows proteins that are similar to the 1zkd. They are 2ex4 and 1im8. These 2 proteins matches 1zkd the most according to DALI. The pictures below shows the structures of the proteins related comparing to 1zkd.
With these informations we can now look into the proteins that is similar to 1zkd and predict the functions. 1im8 is found to be a methyltransferase with a bound S-adenosylhomocysteine from the crystal structure of YecO from Haemophilus influenzae (HI0319). 2ex4is a human methyltransferase with S-adenosylhomocysteine.
Sequence
MIDQTALATEIKRLIKAAGPMPVWRYMELCLGHPEHGYYVTRDPLGREGDFTTSPEISQMFGELLGLWSASVWKAADEPQ TLRLIEIGPGRGTMMADALRALRVLPILYQSLSVHLVEINPVLRQKQQTLLAGIRNIHWHDSFEDVPEGPAVILANEYFD VLPIHQAIKRETGWHERVIEIGASGELVFGVAADPIPGFEALLPPLARLSPPGAVFEWRPDTEILKIASRVRDQGGAALI IDYGHLRSDVGDTFQAIASHSYADPLQHPGRADLTAHVDFDALGRAAESIGARAHGPVTQGAFLKRLGIETRALSLMAKA TPQVSEDIAGALQRLTGEGRGAMGSMFKVIGVSDPKIETLVALSDDTDREAERRQGTHGLEHHHHHH
Information on 1zkd
Located on 2p22.2 on human chromosome with 441 aa
Located on 17E3 on mouse chromosome with 436 aa
Alignment with 2ex4 1ZKD:A 24/25 WRYXELCLGHPEHGYYV--TRDPLGREGDFTTSPEISQXFGELLGLWSASVWKAAD-EPQ 2EX4:A 24/7 IEDEKQFYS----KAKTYWKQIPPTVDGMLGGYGHISSIDINSSRKFLQRFLREGPNKTG 1ZKD:A 81/82 TLRLIEIGPGRGTXXADALRALRVLPILYQSLSVHLVEINPVLRQKQQTLLAGI-RNIHW 2EX4:A 80/64 TSCALDCGAGIGRITKRLLLPL--------FREVDMVDITEDFLVQAKTYLGEEGKRVRN 1ZKD:A 140/141 HD-----SFEDVPEGPAVILANEYFDVLPIHQAIKRETGWHERVIEIGASGELVFGVAAD 2EX4:A 132/116 YFCCGLQDFTPEPDSYDVIWIQWVIGHLT------------------------------- 1ZKD:A 195/196 PIPGFEALLPPLARLSPPGAVFEWRP--DTEILKIASRVRDQGGAALIIDYG--HLRSDV 2EX4:A 161/145 ------------------------DQHLAEFLRRCKGSL-RPNGIIVIKDNMAQE----- 1ZKD:A 251/252 GDTFQAIASHSYADPLQHPGRADLTAHV---DFDALGRAAESIGARAHGPVTQG 2EX4:A 191/175 GVILDD---------------VDSSVCRDLDVVRRIICSAG---LSLLAEERQE
Alignment with 1im8 1ZKD:A 59/60 QXFGELLGLWSASVWKAADEPQTLRLIEIGPGRGTXXADALRALRVLPILYQSLSVHLVE 1IM8:A 42/40 SNIITAIGXLAERFV-----TADSNVYDLGCSRGAATLSARRNI-----NQPNVKIIGID 1ZKD:A 119/120 INPVLRQKQQTLLAGI---RNIHWHD--SFEDVPEGPAVILANEYFDVLPIHQAIKRETG 1IM8:A 92/90 NSQPXVERCRQHIAAYHSEIPVEILCNDIRHVEIKNASXVILNFTLQFLP---------- 1ZKD:A 174/175 WHERVIEIGASGELVFGVAADPIPGFEALLPPLARLSPPGAVFEWRP--DTEILKIASRV 1IM8:A 142/140 ---------------------------------------------PEDRIALLTKIYEGL 1ZKD:A 232/233 RDQG--GAALIIDYG 1IM8:A 157/155 ---NPNGVLVLSEKF
Most probably a methyltransferase as most of the matches from DALI are transferase
S-ADENOSYL-L-HOMOCYSTEINE - Structure that Identify Transferase
DALI Results
SUMMARY: PDB/chain identifiers and structural alignment statistics
NR. STRID1 STRID2 Z RMSD LALI LSEQ2 %IDE REVERS PERMUT NFRAG TOPO PROTEIN 1: 3027-A 1zkd-A 56.8 0.0 349 349 100 0 0 1 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION duf185 (rhodops 2: 3027-A 2ex4-A 11.7 3.0 185 221 12 0 0 22 S TRANSFERASE adrenal gland protein ad-003 (homo sapien 3: 3027-A 1im8-A 11.6 3.2 178 225 10 0 0 18 S TRANSFERASE yeco (methyltransferase, hypothetical pro 4: 3027-A 2gb4-A 10.8 3.3 184 231 13 0 0 19 S TRANSFERASE thiopurine s-methyltransferase (thiopurine 5: 3027-A 2fk7-A 10.7 3.8 186 277 14 0 0 19 S TRANSFERASE methoxy mycolic acid synthase 4 (mycobact 6: 3027-A 2ob1-A 10.2 3.9 196 319 9 0 0 23 S TRANSFERASE leucine carboxyl methyltransferase 1 (prot 7: 3027-A 2f8l-A 10.0 3.7 182 318 8 0 0 22 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION hypothetical pro 8: 3027-A 2avn-A 10.0 3.7 183 242 11 0 0 20 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION ubiquinoneMENAQU 9: 3027-A 2bzg-A 9.9 3.4 182 226 13 0 0 20 S TRANSFERASE thiopurine s-methyltransferase (thiopurine 10: 3027-A 2aot-A 9.8 4.3 182 285 13 0 0 23 S TRANSFERASE histamine n-methyltransferase (hmt) (homo 11: 3027-A 2p8j-A 9.7 3.4 168 203 10 0 0 16 S TRANSFERASE s-adenosylmethionine-dependent methyltrans 12: 3027-A 1vl5-A 9.7 3.3 167 225 9 0 0 18 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION unknown conserve 13: 3027-A 1hnn-A 9.7 3.4 183 261 13 0 0 18 S TRANSFERASE phenylethanolamine n-methyltransferase (p 14: 3027-A 1m6e-X 9.6 3.6 206 359 9 0 0 27 S TRANSFERASE s-adenosyl-l-methionnine:salicylic acid ca 15: 3027-A 2fay-A 9.5 3.9 178 274 14 0 0 21 S 16: 3027-A 2h00-A 9.2 2.9 158 225 9 0 0 18 S TRANSFERASE methyltransferase 10 domain containing pro 17: 3027-A 1wy7-A 9.2 3.1 155 195 13 0 0 15 S TRANSFERASE hypothetical protein ph1948 (pyrococcus h 18: 3027-A 1m6y-A 8.8 4.3 156 289 10 0 0 19 S TRANSFERASE s-adenosyl-methyltransferase mraw (thermo 19: 3027-A 1i9g-A 8.7 3.0 149 264 9 0 0 15 S TRANSFERASE hypothetical protein rv2118c (mycobacter 20: 3027-A 1zq9-A 8.6 3.2 144 278 16 0 0 19 S TRANSFERASE probable dimethyladenosine transferase (s-
Pfam Results
DUF185: domain 1 of 1, from 64 to 299: score 227.1, E = 3.9e-65
*->alArwllveykllgyPYadlnlvElGaGrGtaielmsdlLryiarlv
+l++w + ++k+ ++P l+l E+G+GrGt +m+d+Lr+ r+
query 64 LLGLWSASVWKAADEP-QTLRLIEIGPGRGT---MMADALRA-LRVL 105
PdvyartryylvEiSprLaarQketLapkvaplGhdskveieatdlsglv
P +y+ ++++lvEi+p L+++Q++ La ++ ++
query 106 PILYQSLSVHLVEINPVLRQKQQTLLA-----------------GIR-NI 137
rWhdasileedPdgvptvliaNEVlDalPHDlvrfdkrgggwyErhVlvd
Whd s +e++P+g p v++aNE +D lP +++ +kr+ gw+Er V ++
query 138 HWHD-S-FEDVPEG-PAVILANEYFDVLP--IHQAIKRETGWHER-V-IE 180
ldgdfrlvysqeldplaglaltlreaaldPVKstkklvpsalskllpkll
+ ++lv+++++dp g+ ++l
query 181 IGASGELVFGVAADPIPGFEAL------------------------LPPL 206
ppaeevgygtEvYsParllellqalaerLpahrGrlLaiDYGhlaseyyh
+ +g+++E+ P e+l+++ + + +G++L+iDYGhl+s
query 207 ARLSPPGAVFEW-RPDT--EILKIASRVRD-QGGAALIIDYGHLRSD--- 249
prrksalaaemfngtllqayrqhahddpltnpssllVlyStvaqGlaDiT
g+++qa+ h + dpl +p G+aD+T
query 250 ------------VGDTFQAIASHSYADPLQHP------------GRADLT 275
ahVDFtalaradqyqtaakaagdlkvlgvet<-*
ahVDF+al +aa +g + + g+ t
query 276 AHVDFDALG------RAAESIG-ARAHGPVT 299