Structure of 2ob5: Difference between revisions
mNo edit summary |
mNo edit summary |
||
| Line 3: | Line 3: | ||
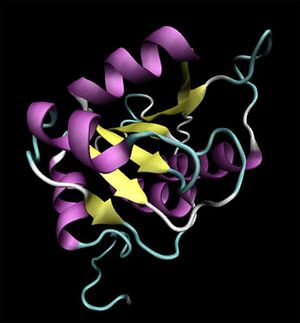
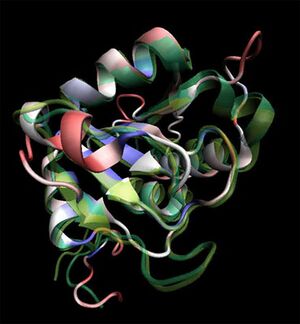
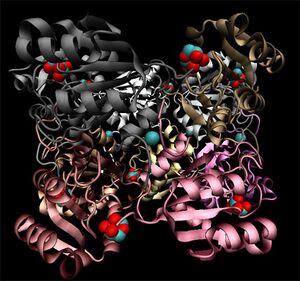
[[Image:2ob5_main.jpg|left|thumbnail|Figure 1. Structure of 2ob5]] | |||
==Crystal structure== | ==Crystal structure== | ||
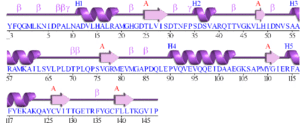
Initial structural analyses indicate that 2ob5A is a member of the RbsD/FucU protein superfamily, and is of the RbsD-like fold classification. It is a globular protein composed of five alpha-helixes arranged around six internal beta strands in the tri-layered a/b/a fold characteristic of RbsD-like proteins. | Initial structural analyses indicate that 2ob5A is a member of the RbsD/FucU protein superfamily, and is of the RbsD-like fold classification. It is a globular protein composed of five alpha-helixes arranged around six internal beta strands in the tri-layered a/b/a fold characteristic of RbsD-like proteins. | ||
Revision as of 22:17, 15 June 2009
Crystal structure
Initial structural analyses indicate that 2ob5A is a member of the RbsD/FucU protein superfamily, and is of the RbsD-like fold classification. It is a globular protein composed of five alpha-helixes arranged around six internal beta strands in the tri-layered a/b/a fold characteristic of RbsD-like proteins.
Structural alignment searches
Dali search
Early structural alignments (such as Dali) suppoorted this classification, returning ribose-transport proteins as high similarity hits. Of the structures found, the majority of high-significance (z > 15) matches were variations on two prokaryotic RbsD proteins, 3e7n (D-ribose high affinity transport system, S. Typhi) and 1ogD (high affinity ribose transporting protein, B. Subtillis). These two crystal structures are shown at right.
Structures of matching proteins
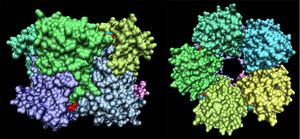
Crystallographic visualisation of these two proteins has revealed that the biological unit of each is a homo-decameric oligomer, which has a ring--like quaternary structure. The complete oligomer of 3e7n is shown below. PQS analysis of both structures suggests that this decameric assembly is correct for each.
Note the ring-like structure of the oligomer, which contains distinct upper and lower pentameric sub-rings which are rotated slightly relative to each other, such that no more than three subunits coincide at any one junction.
Alignment visualisation
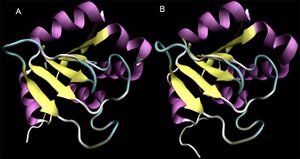
Structural similarity between the monomers of both proteins and 2ob5A is high, despite sequence identity values of less than 20%. To confirm the high structural identity of the Dali results (z=17.9 and 16.9 for 3e7n-c and 1ogd-D, respectively), the proteins were aligned using MultiSeq for VMD and overlaid.
From visual analysis the strength of the alignment is clear.
At left, 2ob5 is coloured by a BLOSUM60 plot of residue conservation. The prevalence of blue residues in the internal beta-sheets indicates high sequence conservation in those regions. This is supported by the molecule's RbsD-like fold classification, as the 6-strand beta-sheet structure is highly conserved in RbsD proteins.
Due to this considerable structural similarity, we conducted an in depth comparison of 2ob5A to the better characterised 3e7n and 1ogd.
Ligand binding site
1ogD and 3e7n
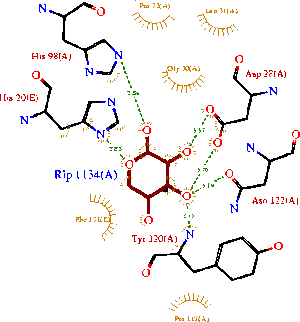
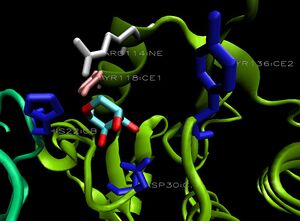
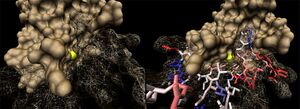
The binding sites of 1ogd are known, and the protein has been crystallised bound to its ribose-D ligand in a pentameric ring quaternary structure. The oligomerisation of the protein appears necessary for the binding of this ligand, as the active site lies in a cleft between proteins adjacent on the same ring
, and includes coordinating residues from both subunits. A LIGPLOT analysis of the 1ogd ribose-D binding site is shown at left.
His20 is donated by the adjacent monomer. This residue arrangement is shown in 3D in at right.
Residues are again coloured by sequence conservation. All five residues are conserved in 3e7n, further supporting their involvement in binding of ribose-D.
Target protein
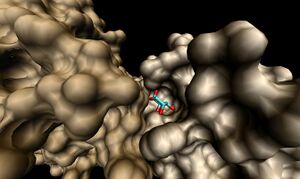
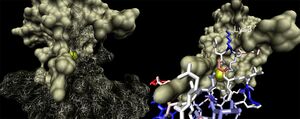
2ob5A was not crystallised with ligand present. As such, it is uncertain whether it too is a ribose binding protein. His20, Tyr120 and Asp28 on 1ogd are all conserved in 2ob5A. Asn122 appears to have undergone a deletion judging from the sequence alignment, and His98 and Lys102 have been substituted for by Arg and Tyr residues, respectively. These differences may suggest a slightly different specificity. However, the 3D Multiseq alignment of 2ob5 with 1ogD and 3e7n retained the binding amino acids in position. Superimposition of the 1ogD ligand onto an aligned 2ob5A suggests that the binding mode and/or ligand of 2ob5 is similar
. Visualisation of the corresponding surfaces indicate that a binding cleft is still present on 2ob5A.
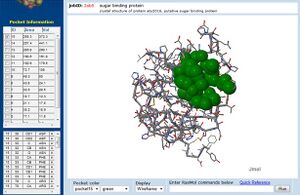
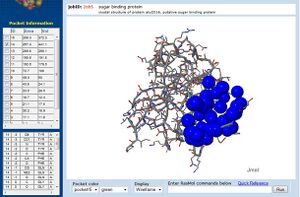
In addition, CASTp analysis predicts the monomer's highest surface-area cleft at this site.
A large pocket is identified by CASTp in 1ogd atthe same site, although this protein's pentameric crystallisation allows its predicted cleft to include the adjacent His20 residue.
Ion-coordination site
1ogD and 3e7n
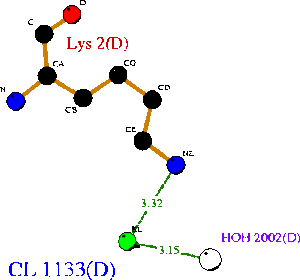
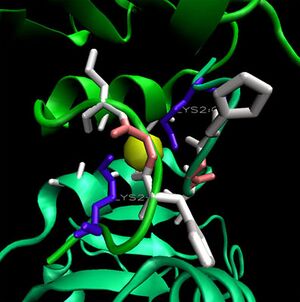
1ogd was cocrystallised with an ion, tentatively labelled as a chloride in this case. The ion coordinating site lies at the amino-terminus of the protein. Ligplot analysis gives Lys2 as a key residue for ion coordination.
Figure X
shows the 3D alignment of two 1ogD pentamers onto the 3e7n decameric scaffold. The chloride ion is visible in the centre of the facing side. Bound Ribose-D molecules are also visible. From this, it is suggested that ions are coordinated by two adjacent monomers; specificlly by the residue Lys 2. Unlike the ribose binding site, however, the ion binding cleft is formed by subunits on different pentameric rings; one from the upper and one lower. The Lys2 residues combine with hydrophobic residues on each subunit's amini-terminal loop to form an ion cage structure.
This is clearly visible in the figure at left. Although the individual hydrophobic residues are poorly conserved, the cage structure and coordinating lysines are also present on 3e7n. It is possible that this ion contributes to the stability of the decameric ring, existing as it does at the interfaces between top and bottom halves of the oligomer.
Target protein
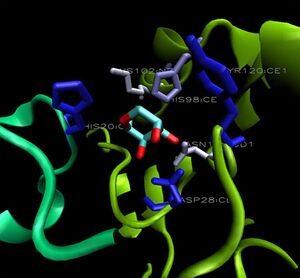
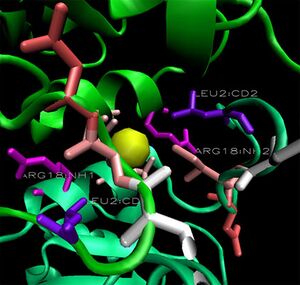
When monomers of 2ob5 are superimposed on the ring structure of 1ogd or 3e7n, the target protein appears to retain its ion cage structure.
However, the critical coordinating lysine is not present. Lys2 has been replaced with a hydrophobic leucine residue (figure at right), which cannot coordinate a Cl ion. The only nearby positive residue - an arginine, which replaces Ala16 on 1ogd - appears in the wrong orientation, although this may be an artifact of the alignment. It is possible that 2ob5A does not have the ability to coordinate an ion at this site.
Surface representations of the 1ogD Cl-binding cleft indicate a buried coordination site. This corresponds to the predicted ion cage structure. The interface between the two subunits covers a comparatively large surface area at this site, and the region as a whole is comparatively well conserved. Of particular note is the residue Lys3, which is conserved across all three crystallised proteins and lies in an extended cleft in the adjacent monomer. This residue and its cleft is conserved in 2ob5, although individual sequence conservation at the cleft is not high.
This, along with a number of corresponding cleft-ligand-like features at the subunit interface of 2ob5, suggests that 2ob5 exists in the same decameric conformation as 1ogd and 3e7n despite the potential lack of ion coordination. Cast-P analysis supports this, as the molecule's second-largest predicted binding surface lies around the putative ion-coordinating region.
Sequence conservation
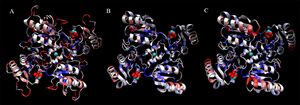
Finally, shown here are representations of a four-subunit face of each protein, demonstrating potential binding sites for D-ribose and Cl. Residues are coloured by sequence conservation between these three proteins.
It is again clear that the structural alignment is strong, even at the quaternary level. Additionally, it can be seen that although 2ob5 has many areas of low conservation, these generally correspond with variable loop regions, and that the key secondary-structural elements remain remarkably constant.