Structure work done by Michael: Difference between revisions
No edit summary |
No edit summary |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
Here is link to Dali file for structural allignment of Fascin 1: | Here is link to Dali file for structural allignment of Fascin 1: | ||
http://ekhidna.biocenter.helsinki.fi/dali_server/results/ | http://ekhidna.biocenter.helsinki.fi/dali_server/results/20090602-0024-75e94ac980c3ad7a77bfb91e13a9d591/index.html | ||
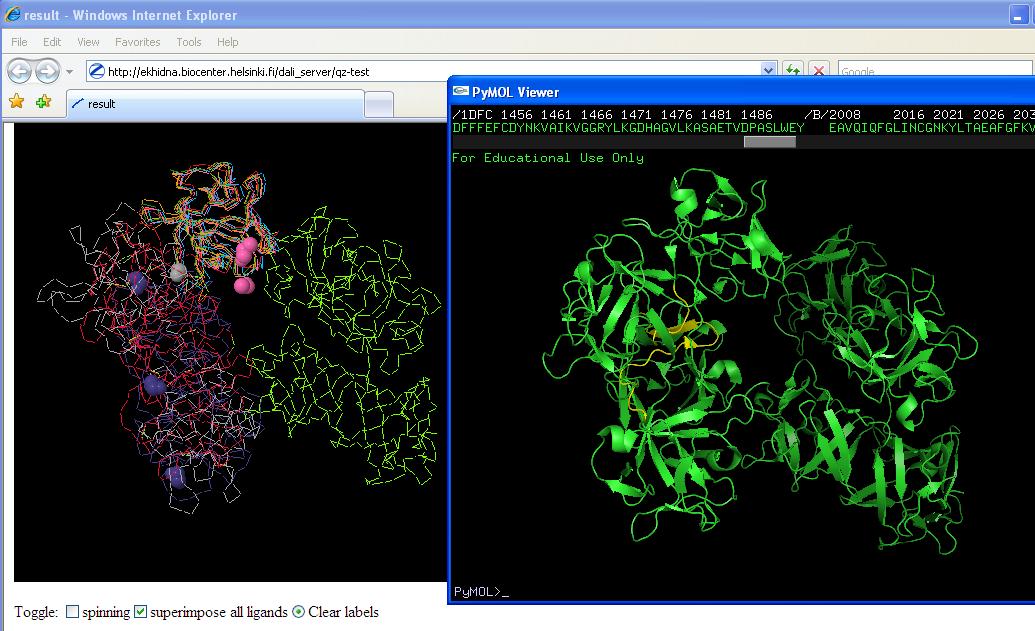
From here comparative structure allignments were possible. All of which were very similar to fascin 1 providing more clues as to function of the protein. | From here comparative structure allignments were possible. All of which were very similar to fascin 1 providing more clues as to function of the protein. | ||
Yellow region on right hand picture is conserved region from sequence multiple alignment. Figure on left is structure allignments (note pink ligands in first and purple ligands in second). | Yellow region on right hand picture is conserved region from sequence multiple alignment. Figure on left is structure allignments (note pink ligands in first and purple ligands in second). | ||
Putative conserved residues are: IAMHPQV (aa's 134-142) and LINRPIIVFRGEHGVIGCRK (aa's 380-400) | |||
List of structures is: mol1A/1dfcA/1jlyA/1ijtA/1qxmB/1ev2A/1bfgA/1axmA/4fgfA/ | List of structures is: mol1A/1dfcA/1jlyA/1ijtA/1qxmB/1ev2A/1bfgA/1axmA/4fgfA/ | ||
| Line 17: | Line 19: | ||
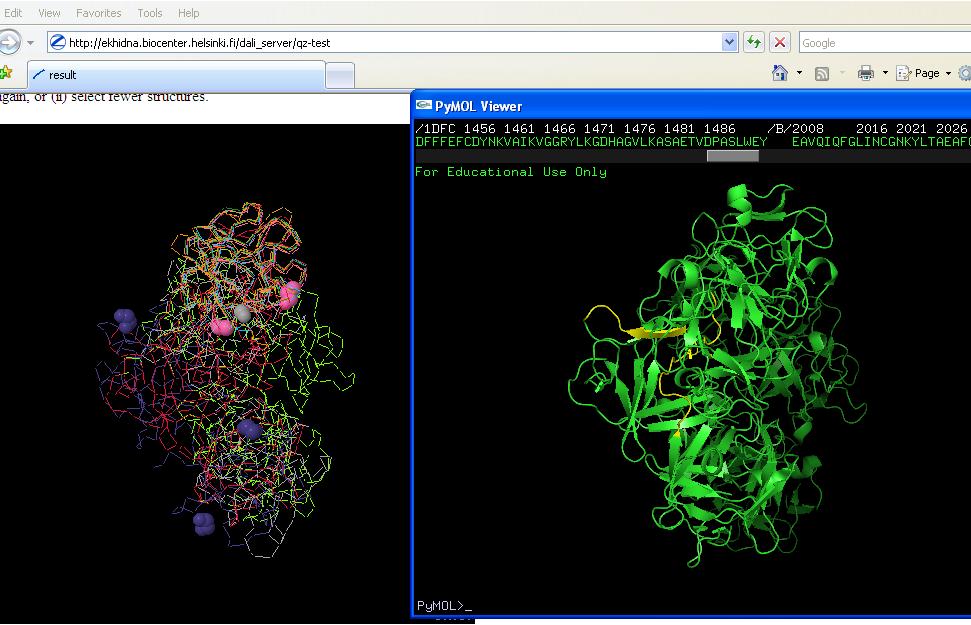
[[Image:Comparison stuff 2.jpeg]] | [[Image:Comparison stuff 2.jpeg]] | ||
cropped print screen of proteins. Purple is: 1qxmB | cropped print screen of proteins. Purple is: 1qxmB | ||
Latest revision as of 07:54, 2 June 2009
Unfortunately PDB databank down so very little work was able to be done.
Here is link to Dali file for structural allignment of Fascin 1: http://ekhidna.biocenter.helsinki.fi/dali_server/results/20090602-0024-75e94ac980c3ad7a77bfb91e13a9d591/index.html
From here comparative structure allignments were possible. All of which were very similar to fascin 1 providing more clues as to function of the protein.
Yellow region on right hand picture is conserved region from sequence multiple alignment. Figure on left is structure allignments (note pink ligands in first and purple ligands in second).
Putative conserved residues are: IAMHPQV (aa's 134-142) and LINRPIIVFRGEHGVIGCRK (aa's 380-400)
List of structures is: mol1A/1dfcA/1jlyA/1ijtA/1qxmB/1ev2A/1bfgA/1axmA/4fgfA/