Surface Properties: Difference between revisions
From MDWiki
Jump to navigationJump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
<font size = "4">''' | <font size = "4">'''Surface Properties'''</font> | ||
[[Image: | [[Image:Document7_07.png]] | ||
'''Figure 13. (A) '''Molecular structure of 2gfh with the ligand PO<sub>4.</sub>''' (B) '''Molecular and chemical structure of PO<sub>4.</sub>''' (C) '''Ligand interaction involving PO<sub>4.</sub> (Picture adapted from Profunc) | |||
the | * Identify the likely biochemical function from the 3D structure | ||
* Possible binding sites and potential ligands - PO<sub>4</sub> | |||
* PO<sub>4</sub> most likely be an active site and function | |||
[[Posphastase|Next Page]] | |||
[[ | |||
Revision as of 23:21, 10 June 2007
Surface Properties
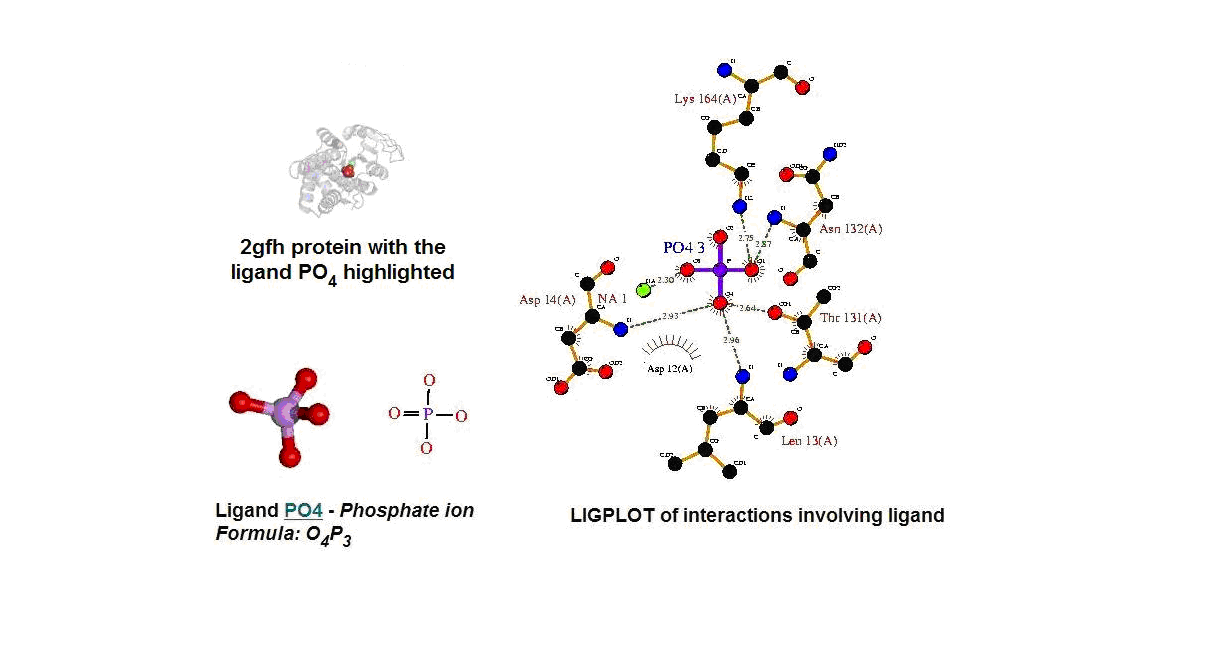 Figure 13. (A) Molecular structure of 2gfh with the ligand PO4. (B) Molecular and chemical structure of PO4. (C) Ligand interaction involving PO4. (Picture adapted from Profunc)
Figure 13. (A) Molecular structure of 2gfh with the ligand PO4. (B) Molecular and chemical structure of PO4. (C) Ligand interaction involving PO4. (Picture adapted from Profunc)
- Identify the likely biochemical function from the 3D structure
- Possible binding sites and potential ligands - PO4
- PO4 most likely be an active site and function