Pyridoxal Phosphatase structure: Difference between revisions
From MDWiki
Jump to navigationJump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
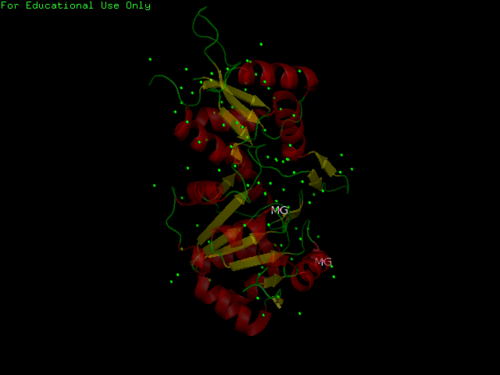
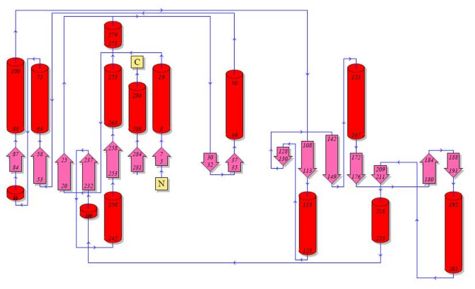
[[Image:2cftA Structure breakdown.jpg|left|thumb|500px|Fig 1. Courtesy (and edited) of Almo et al., 2007. Structure of 2cftA - Human Pyridoxal 5'-Phosphate Phosphatase with its substrate. As mentioned in the "Introduction" section, the core domain consists of beta sheets sandwiched by alpha helices. 2cftA is similar to 2cfsA, with the differences being the presence of a PLP ligand and the type of metal ions (2cfsA has magnesium ions, 2cftA has calcium ions). Notice the similarities between the structure of 2cftA and the PyMOL-generated image of 2cfsA (Fig 2.).]] | |||
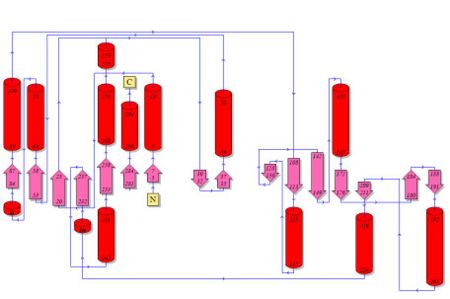
[[Image:2cftA Structure breakdown.jpg|left|thumb|500px|Courtesy (and edited) of Almo et al., 2007. | [[Image:2cfsA_PyMOL.png|left|thumb|500px|Fig 2. The three-dimensional structure of 2cfsA, as generated by PyMOL<BR> | ||
<BR> | <BR> | ||
Red - Helix<BR> | Red - Helix<BR> | ||
Yellow - Sheet<BR> | Yellow - Sheet<BR> | ||
Green - Loop <BR>]] | Green - Loop <BR> | ||
Green Dots - Water molecules]] | |||
<BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR> | <BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR> | ||
'''DALI - 1st Run''' | '''DALI - 1st Run''' | ||
Revision as of 15:19, 7 June 2008

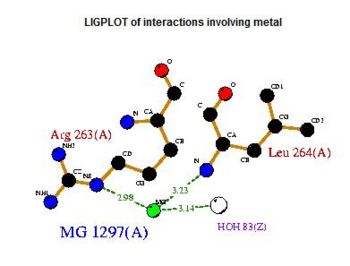
Fig 1. Courtesy (and edited) of Almo et al., 2007. Structure of 2cftA - Human Pyridoxal 5'-Phosphate Phosphatase with its substrate. As mentioned in the "Introduction" section, the core domain consists of beta sheets sandwiched by alpha helices. 2cftA is similar to 2cfsA, with the differences being the presence of a PLP ligand and the type of metal ions (2cfsA has magnesium ions, 2cftA has calcium ions). Notice the similarities between the structure of 2cftA and the PyMOL-generated image of 2cfsA (Fig 2.).
DALI - 1st Run
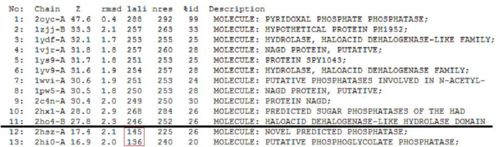
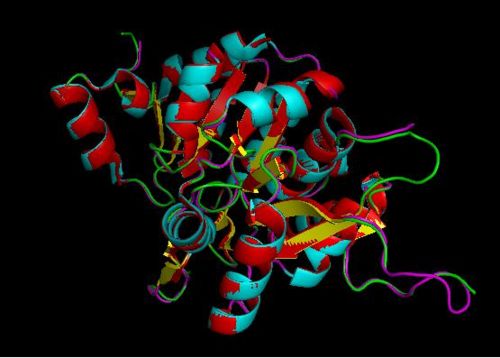
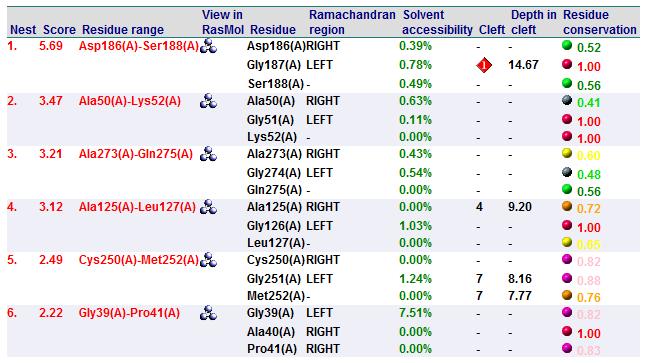
Based on the results of the first run, how similar is 2oycA to 2cfsA? With the aid of PyMOL, 2oycA was super-imposed against 2cfsA...
Comparing 2oycA against 2cfsA
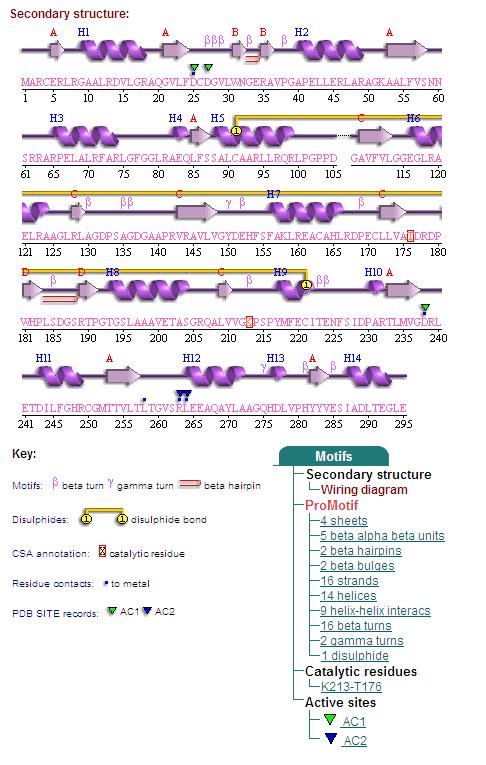
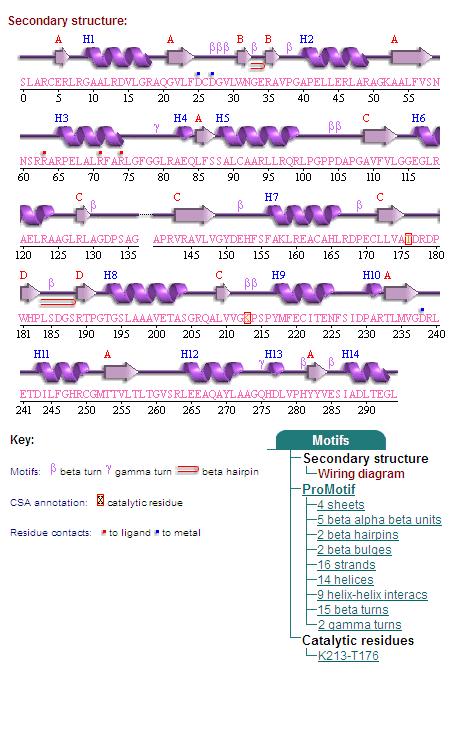
1. Secondary Structure
2. Topology Diagram
PROFUNC
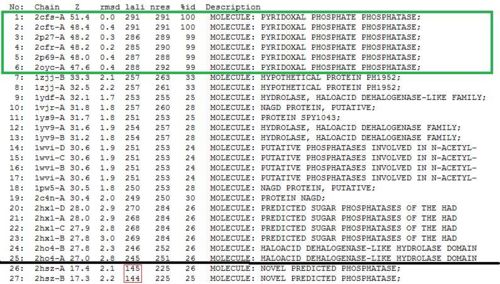
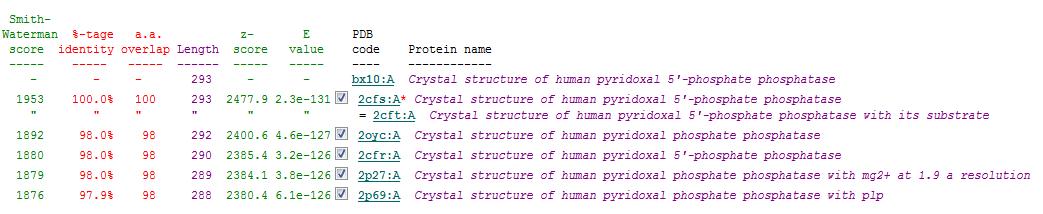
Related Protein Sequences in the PDB (SAS)

Matches to existing PDB Structures
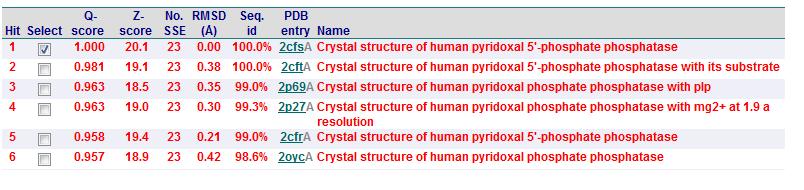
Secondary Structure Matching (SSM)
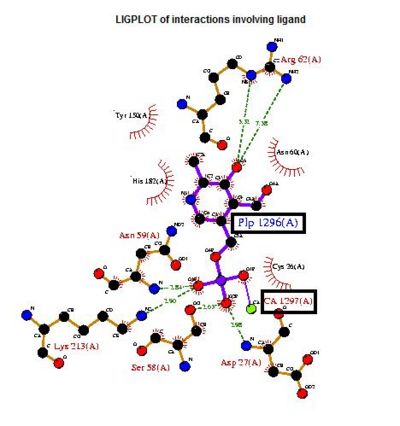
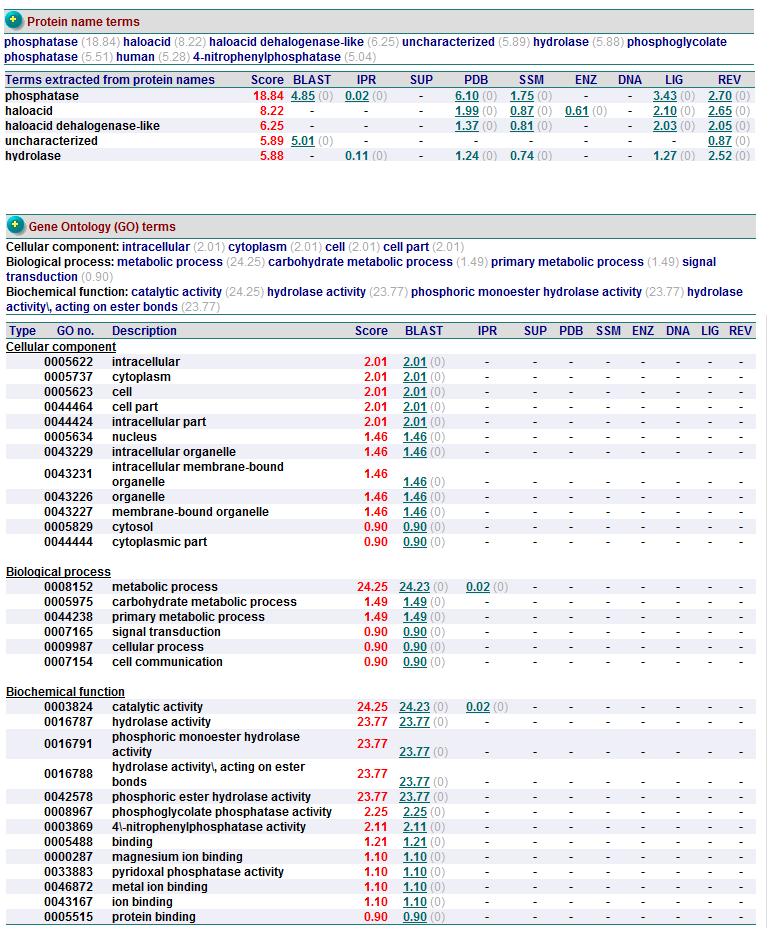
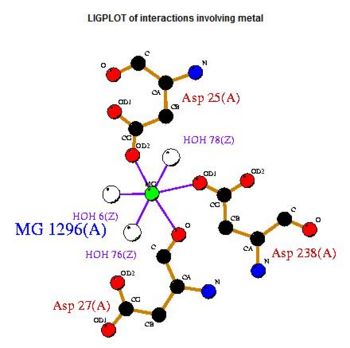
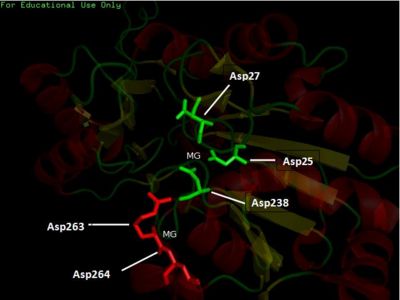
The LIGPLOT of interactions involving the PLP ligand in 2cftA. This was hypothesized to be the location of 2cftA's active site. The Mg 1296(A) ion of 2cfsA is located in the same position as the calcium ion of 2cftA. Therefore, there could be a possibility that the active site of 2cfsA could be at Mg 1296(A).
Dali - 2nd Run
The second DALI run does not affect the results as PROFUNC has already indicated 2cftA to be the most closely-related to 2cfsA. On the contrary, the results serve as a confirmation that 2cfsA is indeed a Pyridoxal Phosphatase and belongs to the same family as 2oycA and 2cftA, 2 proteins that have been studied to aid in the structural and functional prediction of 2cfsA.