Importance of structural studies - the relationship between Evolution, Structure and Function.
- Evolution provides information pertaining to families of structurally related proteins, BUT sequence identities may be low.
- Structural studies help to identify unanticipated relationships in known structures.
- Functions can be assigned based on structural studies.
(Some of the) Results obtained during this study

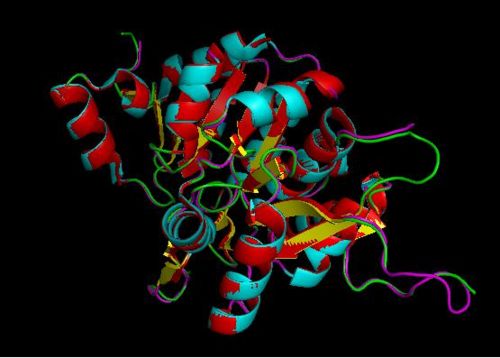
Courtesy (and edited) of Almo et al., 2007. Structure of 2cftA - Human Pyridoxal 5'-Phosphate Phosphatase with its substrate. As mentioned in the "Introduction" section, the core domain consists of beta sheets sandwiched by alpha helices. 2cftA is similar to 2cfsA, with the differences being the presence of a PLP ligand and the type of metal ions (2cfsA has magnesium ions, 2cftA has calcium ions). Notice the similarities between the structure of 2cftA and the PyMOL-generated image of 2cfsA.
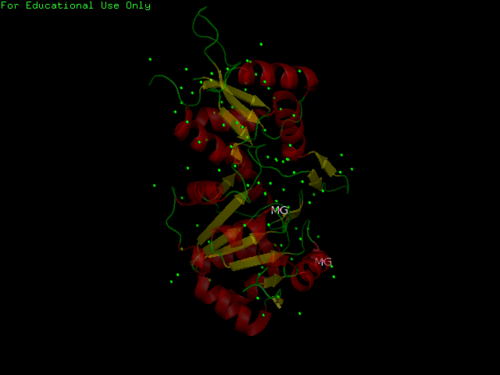
The three-dimensional structure of 2cfsA, as generated by PyMOL
Red - Helix
Yellow - Sheet
Green - Loop
Green Dots - Water molecules
DALI - 1st Run
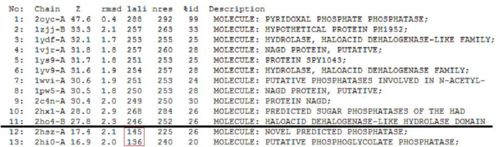
Significant hits as generated from the DALI database
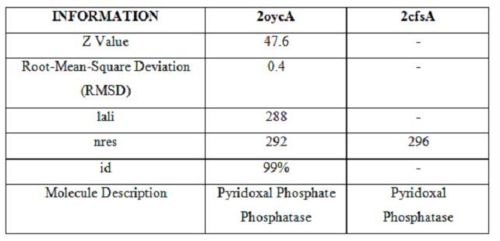
Table 1 - 2oycA, as compared against 2cfsA
Based on the results of the first run, how similar is 2oycA to 2cfsA? With the aid of PyMOL, 2oycA was super-imposed against 2cfsA...
2cfsA vs 2oycA - Similarities
1. Secondary Structure
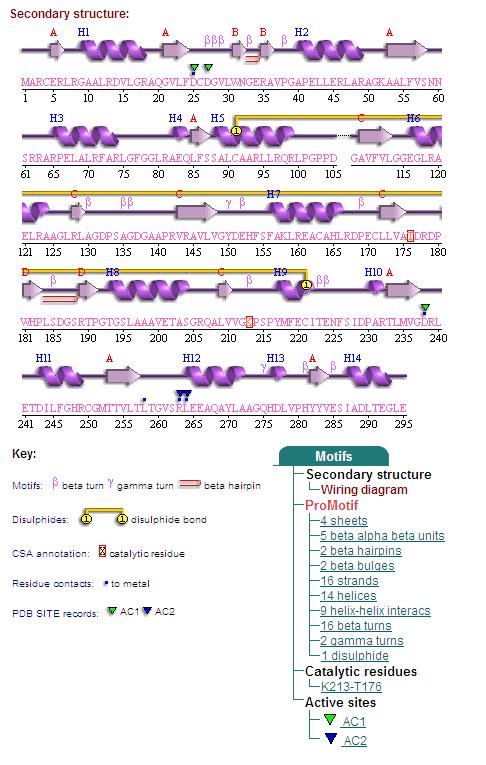
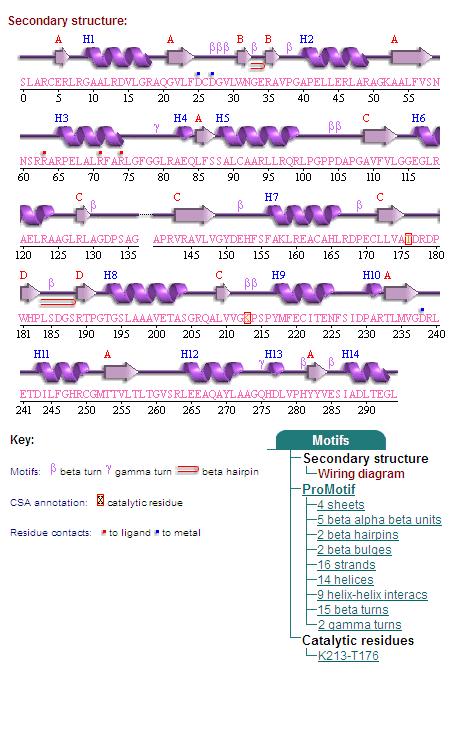
2. Topology Diagram
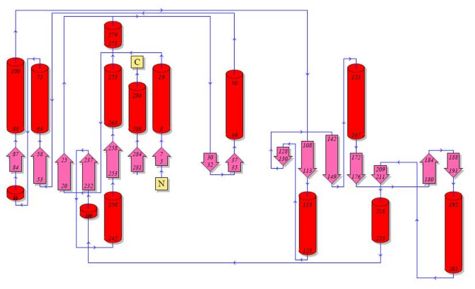
Topology Diagram of 2cfsA
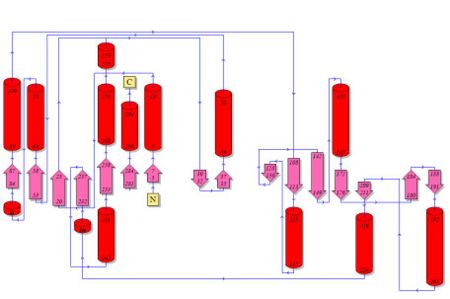
Topology diagram of 2oycA
3. Cleft Analysis (PyMOL)
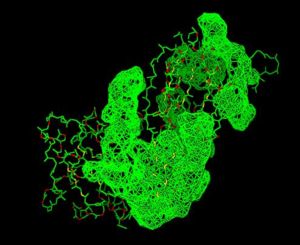
Front View of 2cfsA & its potential binding sites, as generated using PyMOL
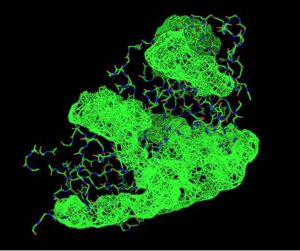
Front View of 2oycA & its potential binding sites, as generated using PyMOL
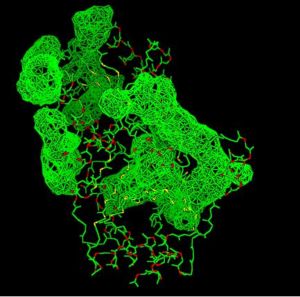
Back View of 2cfsA & its potential binding sites, as generated using PyMOL
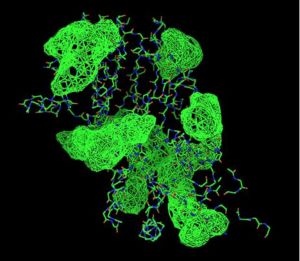
Back View of 2oycA & its potential binding sites, as generated using PyMOL
Pfam
Based on other information obtained from the database, 2cfsA was identified to belong to the Haloacid Dehalogenase (HAD) superfamily. Members of this protein family catalyze phosphoryl transfer of small molecules by co-ordinating the magnesium ions. The substitution of magnesium ions with calcium ions results in a near complete loss of activity. The conclusion, therefore, is that the catalytic site has to be in the region consisting of the magnesium ions.
PROFUNC
Related Protein Sequences in the PDB (SAS)

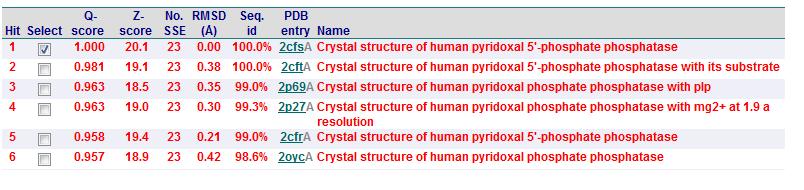
Matches to existing PDB Structures
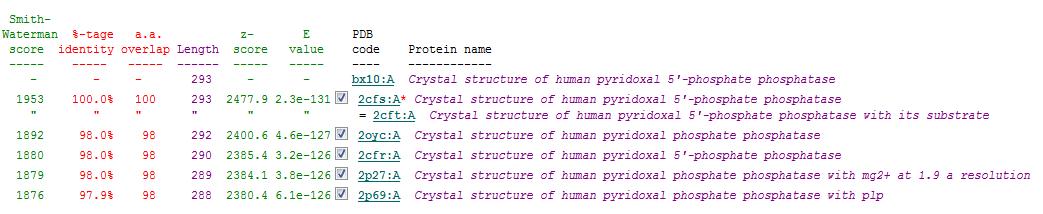
Secondary Structure Matching (SSM)
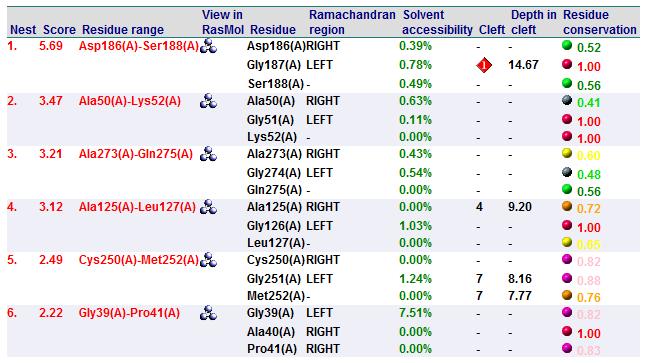
Nest Analysis
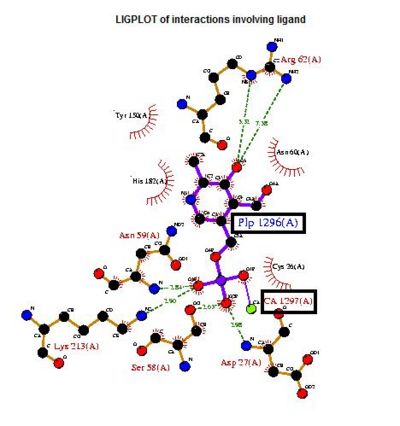
The LIGPLOT of interactions involving the PLP ligand in 2cftA. This was indicated to be the location of 2cftA's active site. The Mg 1296(A) ion of 2cfsA is located in the same position as the calcium ion of 2cftA. Therefore, there could be a possibility that the active site of 2cfsA could be at Mg 1296(A).
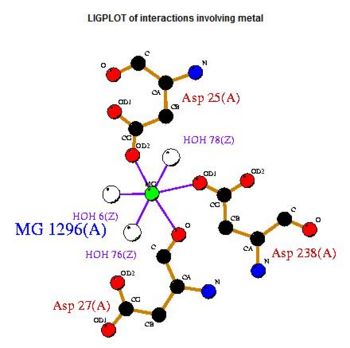
The LIGPLOT of interactions involving the Mg 1296(A) ion in 2cfsA. Notice the similarity of its location with relation to the calcium ion in 2cftA
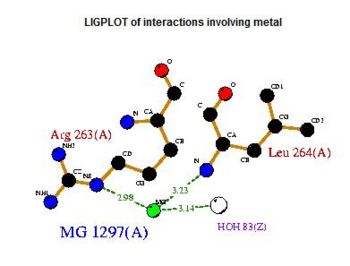
The LIGPLOT of interactions involving the Mg 1297(A) ion in 2cfsA. While it does not seem to share any similarities with the PLP ligand in 2cftA, the information provided by the LIGPLOT will be useful when visualizing the catalytic site of 2cfsA via PyMOL.
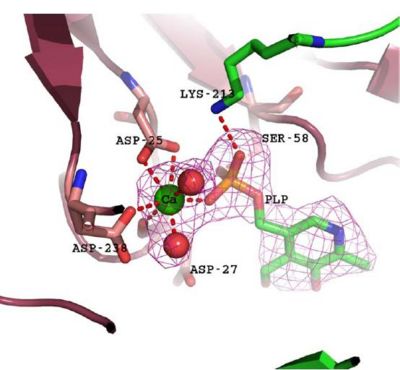
The catalytic site of 2cftA, with its PLP ligand and inhibitory calcium ion. As illustrated, the calcium ion is hepta-coordinated and participates in a bidentate interaction with the active site nucleophile Asp-25, resulting in a loss of activity. Diagram and caption courtesy of Almo et al., 2007.
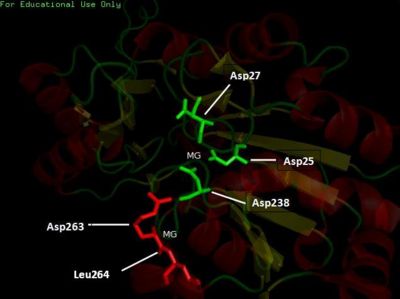
The three-dimensional visualization of the catalytic site of 2cfsA. This was generated based on the location of the residues constituting the catalytic site, as provided in the LIGPLOT diagrams pertaining to the interactions involving 2cfsA's magnesium ions
Based on the above images, the calcium ion (2cftA) is at the same location as one of 2cfsA's magnesium ions (Mg1296). What is the significance?
Substitution of the magnesium ion with the catalytically inert calcium ion results in a near-complete loss of activity.
Dali - 2nd Run
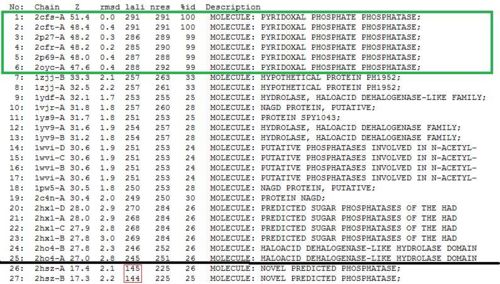
The results obtained when 2cfsA was re-run against the DALI database
"Inconsistencies" - flaw or strength?
The second DALI run does not affect the results as PROFUNC has already indicated 2cftA to be the most closely-related to 2cfsA. On the contrary, the results serve as a confirmation that 2cfsA is indeed a Pyridoxal Phosphatase and belongs to the same family as 2oycA and 2cftA - two proteins that have been studied to assist in the structural and functional prediction of 2cfsA.
Next Up...
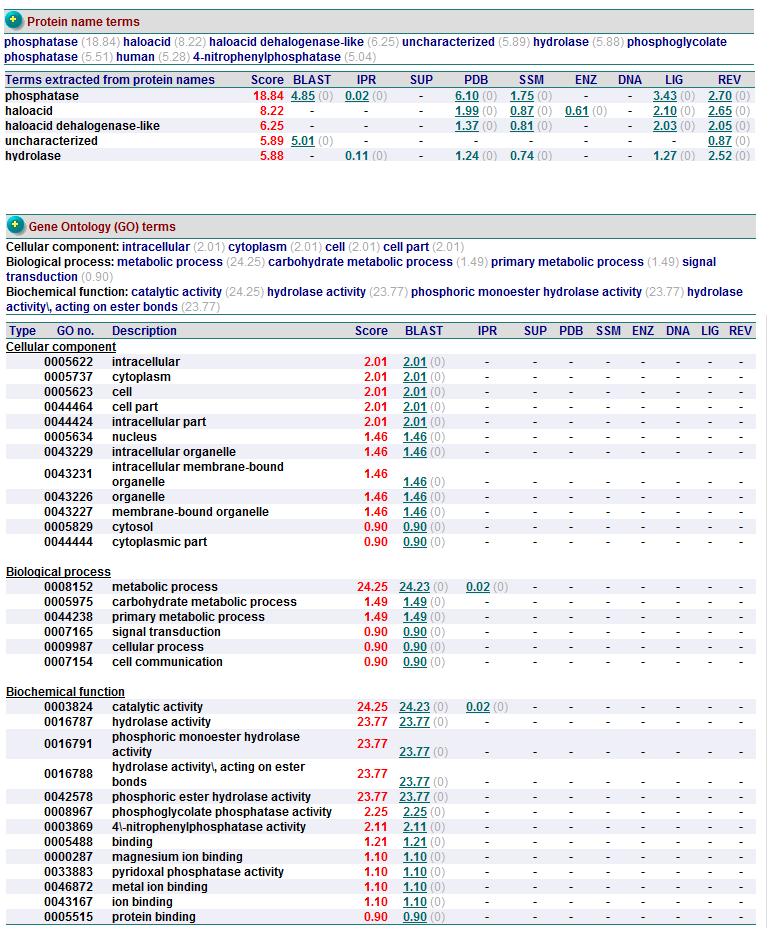
Summary of Predicted Function (ProFunc)
Back To Main Page | Back [Evolution] | Next [Function]