Structure Protein: Difference between revisions
| (10 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
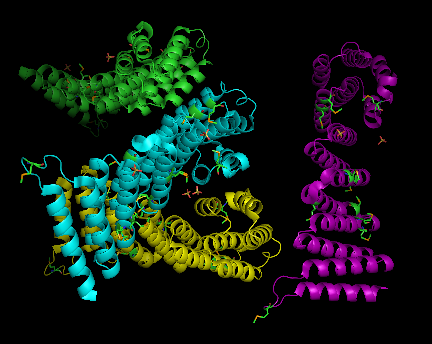
2ifu has 4 chains (A, B, C and D) integrated together to form the whole molecular structure as shown below: | 2ifu has 4 chains (A, B, C and D) integrated together to form the whole molecular structure as shown below: | ||
[[Image:Snap- | [[Image:Snap-gamm ligands.png]] | ||
(chain A= green, chain B= cyan, chain c= purple, and chain d= yellow) | (chain A= green, chain B= cyan, chain c= purple, and chain d= yellow) | ||
Two molecular components were observed: | |||
sulfate ion (SO4)[[Image:Sulfate ion.png]] and selenomethionine (MSE)[[Image:MSE.png]] | |||
Ten sulfate ions were observed. Each ion differs in arrangement as well as orientation related with each chains. | Ten sulfate ions were observed. Each ion differs in arrangement as well as orientation related with each chains. | ||
| Line 16: | Line 17: | ||
== Strutural | == Strutural analysis == | ||
[[SNAP-GAMMA DALI]] | [[SNAP-GAMMA DALI]] | ||
| Line 46: | Line 47: | ||
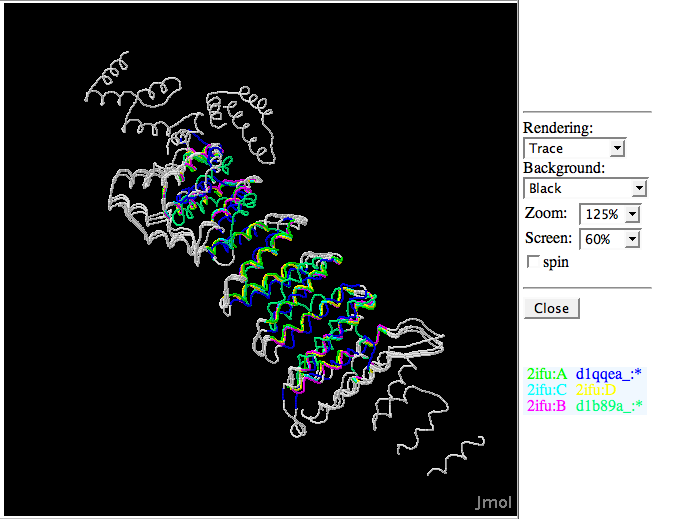
Superimposed structure between 2ifuA; 2ifuB; 2ifuC; 2ifuD; 1qqeA; and 1b89A | Superimposed structure between 2ifuA; 2ifuB; 2ifuC; 2ifuD; 1qqeA; and 1b89A | ||
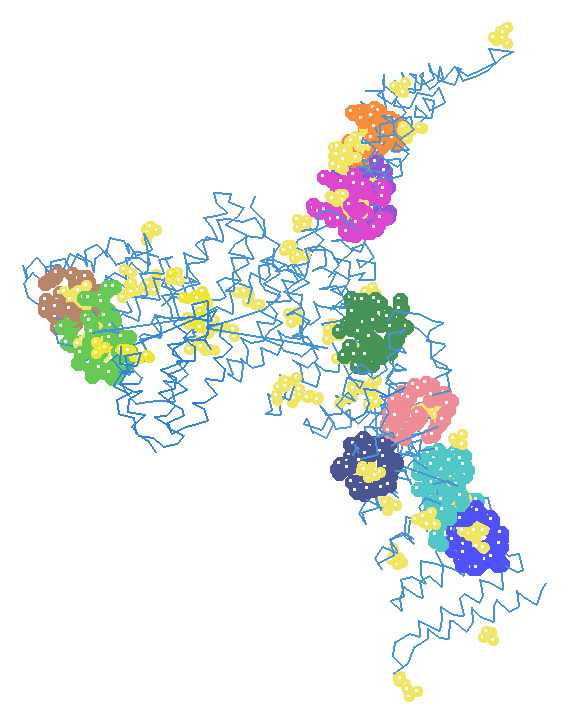
=== ligand binding domain === | |||
[[Image:Ligandbindingsitepredictioni2.png]] | |||
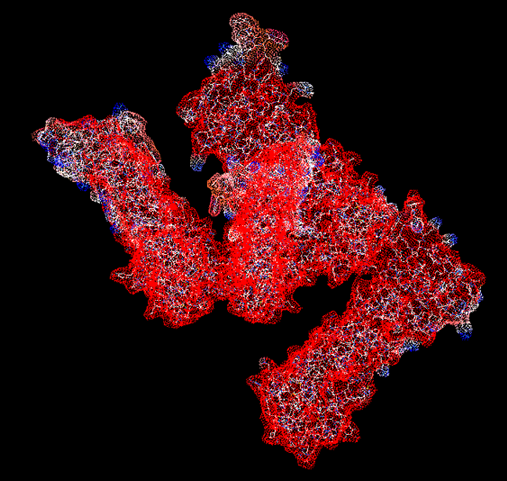
=== electrostatic potential === | |||
[[Image:Electrostatic potential (molecular surface).png]] | |||
electrostatic potential of proteins mapped into molecular surface. | |||
positive potentials are shown in blue while that of negative potential are in red. | |||
the electrostatic potential was calculated using Coulomb computational method. | |||
== Protein families == | == Protein families == | ||
has 2 domains: | has 2 domains: | ||
[http://www.sanger.ac.uk/cgi-bin/Pfam/getblast?id= | [http://www.sanger.ac.uk/cgi-bin/Pfam/getblast?id=204C66wIAJ39p9696N0 Pfam at Sanger] | ||
alignment with: | |||
Pfam-B_7270 (7-66) | |||
highly conserved residue: | |||
Pfam-B_15198 (87-307) | |||
highly conserved residue :residue 104, 109, 121, 168 | highly conserved residue :residue 104, 109, 121, 168 | ||
| Line 65: | Line 86: | ||
[[Image:2ifuA interpro.png]] | [[Image:2ifuA interpro.png]] | ||
== | == links == | ||
CRYSTAL STRUCTURE OF THE VESICULAR TRANSPORT PROTEIN SEC17 | |||
[http://www.pdb.org/pdb/explore/explore.do?structureId=1QQE 1QQE] | [http://www.pdb.org/pdb/explore/explore.do?structureId=1QQE 1QQE] | ||
CLATHRIN HEAVY CHAIN PROXIMAL LEG SEGMENT (BOVINE) | |||
[http://www.rcsb.org/pdb/explore/explore.do?structureId=1B89 1B89] | [http://www.rcsb.org/pdb/explore/explore.do?structureId=1B89 1B89] | ||
[http://www.expasy.org/uniprot/Q5BJK3 uniprot] | |||
http://myhits.isb-sib.ch/cgi-bin/motif_scan | |||
http://www.ncbi.nlm.nih.gov/Structure/vast/vastsrv.cgi?sdid=181604 | |||
http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi | |||
[[Image:ClustalX-alignment.JPG ]] | |||
Latest revision as of 18:52, 9 June 2007
2ifu has 4 chains (A, B, C and D) integrated together to form the whole molecular structure as shown below:
(chain A= green, chain B= cyan, chain c= purple, and chain d= yellow)
Two molecular components were observed:
sulfate ion (SO4)and selenomethionine (MSE)
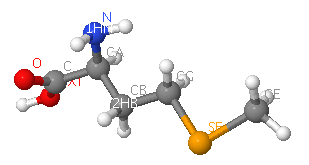
Ten sulfate ions were observed. Each ion differs in arrangement as well as orientation related with each chains.
protein sequence (FASTA)
>gi|118138356|pdb|2IFU|A Chain A, Crystal Structure Of A Gamma-Snap From Danio Rerio AIAAQKISEAHEHIAKAEKYLKTSFXKWKPDYDSAASEYAKAAVAFKNAKQLEQAKDAYLQEAEAHANNR SLFHAAKAFEQAGXXLKDLQRXPEAVQYIEKASVXYVENGTPDTAAXALDRAGKLXEPLDLSKAVHLYQQ AAAVFENEERLRQAAELIGKASRLLVRQQKFDEAAASLQKEKSXYKEXENYPTCYKKCIAQVLVQLHRAD YVAAQKCVRESYSIPGFSGSEDCAALEDLLQAYDEQDEEQLLRVCRSPLVTYXDNDYAKLAISLKVPGGG GGKKKPSASASAQPQEEEDDEYAGGLC
Strutural analysis
Based on Dali search, 6 proteins with highest Z-value were chosen: 1qqe-A (protein transport), 2fi7-A (protein binding), 1a17 (hydrolase), 2ak6-A, 1fch-A (signalling protein), 1haz4-A (transcription factor),
snap-gamma: multiple structure alignment
Snap-gamma: sequence alignment with 1QQE:A
Dark colors: 2ifu:A
Light colors: 1qqe:A
SCOP
multiple alignment results, click here
Superimposed structure between 2ifuA; 2ifuB; 2ifuC; 2ifuD; 1qqeA; and 1b89A
ligand binding domain
electrostatic potential
electrostatic potential of proteins mapped into molecular surface. positive potentials are shown in blue while that of negative potential are in red. the electrostatic potential was calculated using Coulomb computational method.
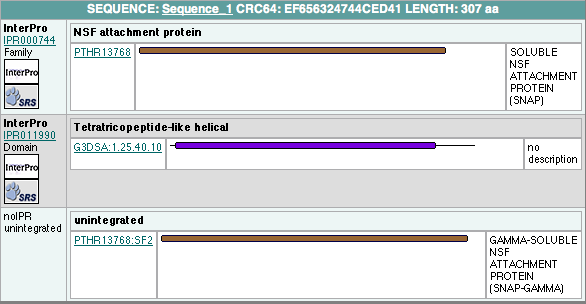
Protein families
has 2 domains: Pfam at Sanger
alignment with:
Pfam-B_7270 (7-66)
highly conserved residue:
Pfam-B_15198 (87-307)
highly conserved residue :residue 104, 109, 121, 168
scorecons result indicated a Diversity of position scores: 89.4% where 100%=high diversity
InterProScan Results
links
CRYSTAL STRUCTURE OF THE VESICULAR TRANSPORT PROTEIN SEC17 1QQE
CLATHRIN HEAVY CHAIN PROXIMAL LEG SEGMENT (BOVINE) 1B89
http://myhits.isb-sib.ch/cgi-bin/motif_scan
http://www.ncbi.nlm.nih.gov/Structure/vast/vastsrv.cgi?sdid=181604