Evolution ERp18: Difference between revisions
No edit summary |
No edit summary |
||
| Line 34: | Line 34: | ||
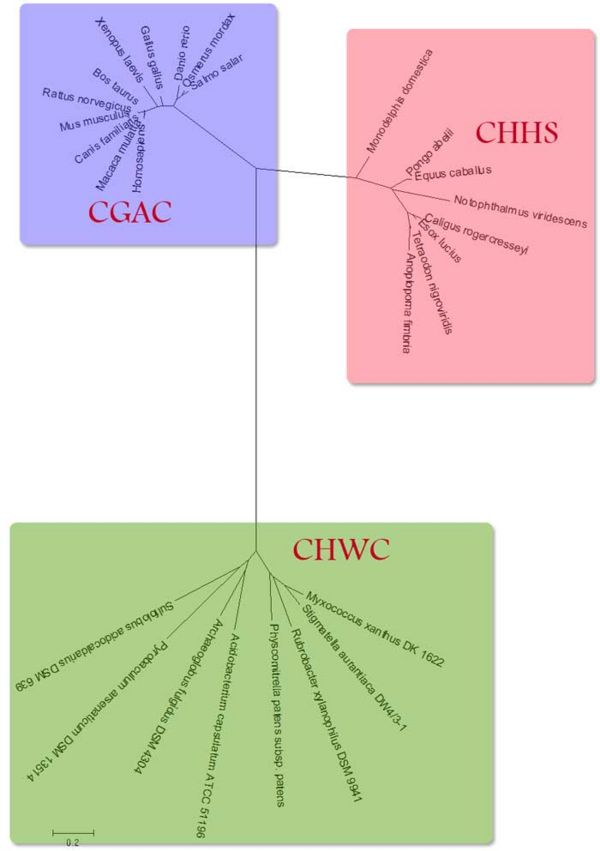
Figure 2 shows in detail the relationship between the three apparent classes of proteins. In both groups of 'higher' organisms the similar trends can be seen where the proteins in more complex organisms are further from the common ancestor. | Figure 2 shows in detail the relationship between the three apparent classes of proteins. In both groups of 'higher' organisms the similar trends can be seen where the proteins in more complex organisms are further from the common ancestor. | ||
[[Image:Tree-domains-cml.jpg|center|thumb|600px|'''Fig. 2'''Unrooted phylogenetic tree showing homologies of different proteins lablled as 'Thioredoxins']] | [[Image:Tree-domains-cml.jpg|center|thumb|600px|'''Fig. 2'''Unrooted phylogenetic tree showing homologies of different proteins lablled as 'Thioredoxins' [1-4]]] | ||
| Line 42: | Line 42: | ||
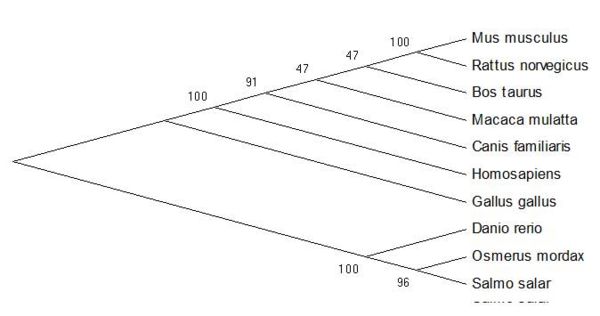
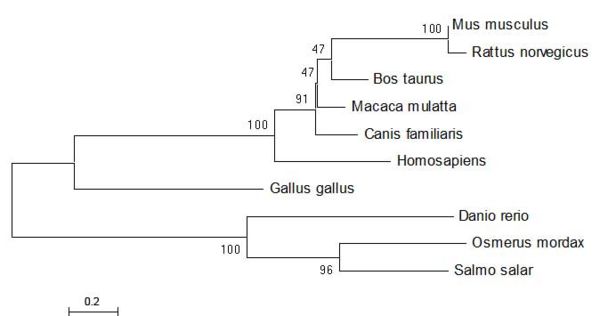
From the sequence alignment, it is seen that the 'CGAC' domain in conserved as is the approxmiate length of the protein sequences. The two following figures show how the ER thioredoxin proteins are related to each other. These trees are 'Neighbour-Joining' Trees and in this circumstance radial, unrooted trees are not appropriate as they add no new information to the analysis. | From the sequence alignment, it is seen that the 'CGAC' domain in conserved as is the approxmiate length of the protein sequences. The two following figures show how the ER thioredoxin proteins are related to each other. These trees are 'Neighbour-Joining' Trees and in this circumstance radial, unrooted trees are not appropriate as they add no new information to the analysis. | ||
[[Image:Tree-CGAConly-cml.jpg|center|thumb|600px|'''Fig. 3''']] | [[Image:Tree-CGAConly-cml.jpg|center|thumb|600px|'''Fig. 3''' [1-4]]] | ||
[[Image:Tree-CGAConly-distance-cml.jpg|center|thumb|600px|'''Fig. 4''']] | [[Image:Tree-CGAConly-distance-cml.jpg|center|thumb|600px|'''Fig. 4''' [1-4]]] | ||
ERp18 seems to be well conserved within the group or organisms and shows little evolutionary distance between species. | ERp18 seems to be well conserved within the group or organisms and shows little evolutionary distance between species. | ||
| Line 52: | Line 52: | ||
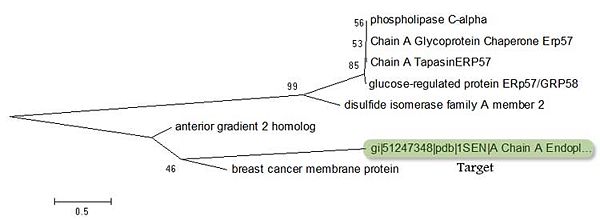
When conducting BLAST searches for the target protein, two other protiens appear as 'related'. These are the 'Breat Cancer Membrane Protein' and the 'Anterior Gradient 2 Homologue'. By comparing these two proteins against other proteins which also have high identity to the target is can be seen these two proteins are related to the target. | When conducting BLAST searches for the target protein, two other protiens appear as 'related'. These are the 'Breat Cancer Membrane Protein' and the 'Anterior Gradient 2 Homologue'. By comparing these two proteins against other proteins which also have high identity to the target is can be seen these two proteins are related to the target. | ||
[[image:Humanproteins-cml.jpg|center|thumb|600px|'''Fig. 5 Evolutionary relationships of 8 taxa ''' | [[image:Humanproteins-cml.jpg|center|thumb|600px|'''Fig. 5 Evolutionary relationships of 8 taxa ''' [1-4]. ]] | ||
There is quite a lot of evolutionary distance between ERp18 and the 'Breat Cancer Membrane Protein' and the 'Anterior Gradient 2 Homologue'. However, it is clear that they have a common ancestor and that ERp18 and the 'Anterior Gradient 2 homologue' have a common ancestor in the 'Breast Cancer Membrane Protein'. It is important to note that the 'Breat Cancer Membrane Protein' and the 'Anterior Gradient 2 Homologue' do not contain the CXXC motif. | There is quite a lot of evolutionary distance between ERp18 and the 'Breat Cancer Membrane Protein' and the 'Anterior Gradient 2 Homologue'. However, it is clear that they have a common ancestor and that ERp18 and the 'Anterior Gradient 2 homologue' have a common ancestor in the 'Breast Cancer Membrane Protein'. It is important to note that the 'Breat Cancer Membrane Protein' and the 'Anterior Gradient 2 Homologue' do not contain the CXXC motif. | ||
| Line 62: | Line 62: | ||
==References== | ==References== | ||
# | #Saitou N & Nei M (1987) '''"The neighbor-joining method: A new method for reconstructing phylogenetic trees."''' Molecular Biology and Evolution 4:406-425. | ||
# | #Schwarz R & Dayhoff M (1979) '''"Matrices for detecting distant relationships."''' In Dayhoff M, editor, Atlas of protein sequences, pages 353 - 58. National Biomedical Research Foundation. | ||
# | #Tamura K, Dudley J, Nei M & Kumar S (2007) '''"MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0."''' Molecular Biology and Evolution 24:1596-1599. | ||
# | #Felsenstein J (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783-791. | ||
#Stephen F. Altschul, Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), '''"Gapped BLAST and PSI-BLAST: a new generation of protein database search programs"''', Nucleic Acids Res. 25:3389-3402. | |||
#Alejandro A. Schäffer, L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001), '''"Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements"''', Nucleic Acids Res. 29:2994-3005. | |||
Revision as of 21:47, 15 June 2009
This page discusses the evolution of the target protein, Endoplasmic reticulum thioredoxin superfamily member.
Methods
To generate a collection of sequences which were, apparently, related to the target protein (ERp18), a PSI-Blast search was conducted. PSI-BLAST is advantageous as it uses an iterative approach whereby selected, relavent results from previous searches are used to inform the next search operation. The PSI-BLAST method was particularly useful in this circumstance since ERp18 is part of a superfamily of proteins and so has many homologues which have high identity scores and low e-vaulues but are actually different proteins.
The collected sequences were analysed using the MEGA4 suite. This program packages allows:
- multiple sequence alignment,
- phylogenetic tree generation,
- bootstrapping, and,
- viewing and editing of phylogenetic trees
All of which were important to this assignment.
The Dayhoff Matrix was used to generate phylogenetic trees throughout this analysis. All bootstrap exercises used 1000 replicates.
Results
Protein Families
The defining motif for a protein in the thioredoxin protein superfamily is the CXXC motif, with the two cystines representing the catalytic residues. It has been suggested in literature the the 'wild card' proteins between the cystines, in part, dictact the specific molecular function (see Function ERp18).
This statement is supported in part, by examining the evolution of proteins which have high identities to the target protein in different organisms. Examining the alignment shows the proteins are varied in the critical CXXC residues however they are likley to be related.
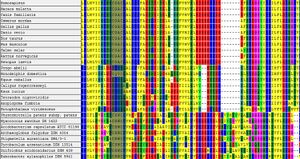
Figure 1 shows the CLUSTAL-W multiple sequence alignment for a selection of proteins which are annotated as 'Thioredoxin' or 'Thioredoxin Like' proteins. Examination of the key functional residues shows that there are at least three classes in the family, containing the CGAC, CHWC, CHHS motifs. CHHS clearly does not conform to the requirement for being a thioredoxin protein as it does not contain the CXXC motif, yet is has the stated annotation. The title 'Thioredoxin-like' may refer to the overall similarity in the sequences rather than the spefic domain.
Another observation from figure 1, is the convervation of the CXXC motif between higher organisms (Homo Sapiens, Salmo Salar, etc) and bacteria and archea. Bacteria and Archea contain the motif 'CHWC', in place of 'CGAC' which is seen in higher organisms. This suggests that the ERp18 protein has evolved from bacteria or archea. It is unlikley that lateral gene transfer has occured, however, it cannot be ruled out based on this evidence.
Figure 2 shows in detail the relationship between the three apparent classes of proteins. In both groups of 'higher' organisms the similar trends can be seen where the proteins in more complex organisms are further from the common ancestor.
Phylogenetic Trees
Although there appear to be many thioredoxin and thioredoxin like proteins, there are very few which have been located in the endoplasmic reticulum. This specific protein, based on the sequences available, appears to be unique to multicellular eukaryotes.
From the sequence alignment, it is seen that the 'CGAC' domain in conserved as is the approxmiate length of the protein sequences. The two following figures show how the ER thioredoxin proteins are related to each other. These trees are 'Neighbour-Joining' Trees and in this circumstance radial, unrooted trees are not appropriate as they add no new information to the analysis.
ERp18 seems to be well conserved within the group or organisms and shows little evolutionary distance between species.
Apparent Human Homologues
When conducting BLAST searches for the target protein, two other protiens appear as 'related'. These are the 'Breat Cancer Membrane Protein' and the 'Anterior Gradient 2 Homologue'. By comparing these two proteins against other proteins which also have high identity to the target is can be seen these two proteins are related to the target.
There is quite a lot of evolutionary distance between ERp18 and the 'Breat Cancer Membrane Protein' and the 'Anterior Gradient 2 Homologue'. However, it is clear that they have a common ancestor and that ERp18 and the 'Anterior Gradient 2 homologue' have a common ancestor in the 'Breast Cancer Membrane Protein'. It is important to note that the 'Breat Cancer Membrane Protein' and the 'Anterior Gradient 2 Homologue' do not contain the CXXC motif.
Discussion
From the analysis and discussion above, it can be suggested that the ERp18 protein is unique to multicelluar organisms and in that group is quite highly conserved. What also may be concluded is that the ERp18 is part of a unique familiy of proteins which belongs to a larger 'superfamily' of thioredoxin proteins and contain the specific CGAC motif.
References
- Saitou N & Nei M (1987) "The neighbor-joining method: A new method for reconstructing phylogenetic trees." Molecular Biology and Evolution 4:406-425.
- Schwarz R & Dayhoff M (1979) "Matrices for detecting distant relationships." In Dayhoff M, editor, Atlas of protein sequences, pages 353 - 58. National Biomedical Research Foundation.
- Tamura K, Dudley J, Nei M & Kumar S (2007) "MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0." Molecular Biology and Evolution 24:1596-1599.
- Felsenstein J (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783-791.
- Stephen F. Altschul, Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402.
- Alejandro A. Schäffer, L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001), "Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements", Nucleic Acids Res. 29:2994-3005.