Function ERp18
Oxidation, reduction, isomerisation activities of ERp18
To date, all members of the thioredoxin superfamily have been found to have the characterising CXXC active site motif. Both cysteine residues are required for oxidative activity and it is the stability of the active site in oxidation states that determines the function of the individual proteins. Thioredoxin-like proteins function as disulfide reductases (Trx), thiol oxidases (DsbA) or di-sulfide isomerases (PDI). (Alanen et al. 2003; Jeong et al. 2008)
The active site motif of the Trx family of proteins is typically CGPC while the PDI family is generally CGHC and the DsbA family is usually CPHC. ERp18 however has an unusual and unique motif of CGAC. Since this is the active site of the protein, it is interesting to note the uniqueness of this domain. Although it is individualistic, it does however help to propose the function of the protein. The CGAC motif of ERp18 is closer to that the Trx and PDI domains than the DsbA domain suggesting that ERp18 could be involved in thiosulfide oxidation or isomerisation of disulfide bonds within the endoplasmic reticulum.
Generally, ERp18 is listed in the databases as a thioredoxin like protein. However, Alanen et al (2003) presented guanidinium chloride denaturation curves, for both the oxidated and reduced forms of the ERp18 protein (fig1a) and PDI domain a (fig 1b), which indicate that ERp18, like the PDI domain a, is more stable in its reduced form. The stability of ERp18 in its reduced form over its oxidation form is a characteristic shared with DsbA and PDI families but not with Trx families. This again suggests that ERp18 is involved more so in the oxidation, or forming of disulfide bonds, than in the reduction.
Inhibition of ER Stress-induced Apoptosis
An accumulation of msifolded proteins can increase cell stress which can ultimately lead to apoptosis of the cell. It has been suggested that ERp18 is involved in reducing stress induced apoptosis.
Jeong et al (2008) demonstrated that that an over expression of the wild type ERp16 protein, the mature form of the ERp18 protein, reduced brefeldin A stress induced apoptosis by approximately 50 percent. The double cystine mutant, ERp16 –CS, replaced both cystine residues located at the active site with serine. It was found that in cells where ERp-16-CS was expressed, apoptosis was increased by a factor of approximately 1.5. This is demonstrated in Figure 2. In absence of ERp16, the potential for cell apoptosis increased.
Location of ERp18 in cells.
ERp18 has been found to be expressed in a number of different cells and organisms. A Serial Analysis of Gene Expression (SAGE) indicated that ERp18 is widely expressed in humans, especially in the brain, prostate, testis, uterus, placenta, kidney, lung, liver and heart. (Alanen et al. 2003; Liu et al. 2003; Jeong et al. 2008). Lash et al (2000) and Ye et al (2002) give a more detailed method of the SAGE analytical method.
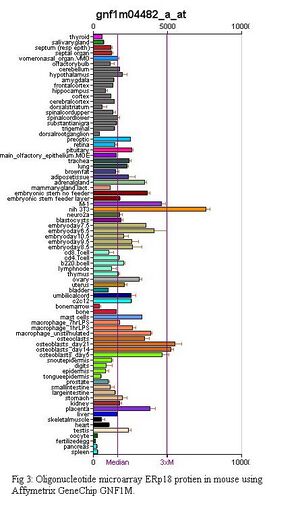
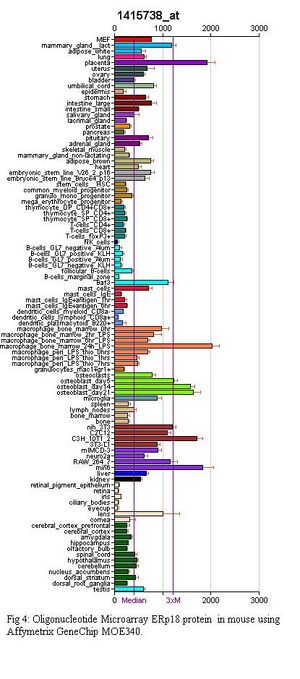
In mice, an oligonucleotide arrays were conducted using Affymetrix GeneChip microarray; GeneAtlas GNF1M and GeneAtlas MOE430. More information on the development of these can be found in the paper by Su et al. (2004).
Figure 3 shows the results of the GNF1M array while Figure 4 shows the results for the Gene Atlas MOE340 array. From these experiments it can be seen that ERp18 is highly expressed in bone marrow macrophages which have been induced with bacterial Lipposacharides (LPS), placenta, the pancreatic beta cell line for mice (Min6), mouse embryo cell line (C3h_10t1_1), osteoblasts and the mouse fibroblast cell line(Nih 3T3).
This indicates that ERp18 is possibly important in the early stages of organism development and also helps to support the theory that ERp18 is involved in inhibiting stress induced apoptosis as it is located in cell lines where misfolded proteins and apoptosis of cells can be detrimental to the health of the organism.
Analytical Methods
Some discrepancy can be seen between the MOE 430 and GNF1M microarray results even though the microarray chips are products of Affymetrix and are analysed using GeneChip Robust Multiarray Average (GCRMA) methods. The discrepancy is most likely a result of different mice, at different developmental stages being sampled from. GeneAtlas MOE430 chips were created using male mice between 8-10 weeks old. Female mice were sampled for female only cell lines.
The difference in expression between the humans and the mice cell lines could be a result of a number of different factors including the different analytical methods. The SAGE method does not require prior knowledge of the sequence of gene transcription and therefore does not create limitations because of genome annotation and gene prediction. SAGE methods tend to be more accurate because not only can they accurately determine the absolute abundance of mRNA but they can also detect any slight differences in expression levels between cells. As such, SAGE methods tend to be better for the identification of new genes.(Lash et al. 2000; Ye et al. 2002). Another reason for the discrepancy can be due to the difference in the human and mice and can therefore highlight the idea that mice might not be the most reliable model for this particular protein.
Functional Summary
Structurally, it is clear that ERp18 is a member to the thioredoxin superfamily. However the unique catalytic motif means that it is independent of all three thioredoxin like protein families. It has been found that ERp18 is more stable in its reduced state suggesting that it is involved in the formation, not breaking of disulfide bonds. It is also proposed that ERp18 is involved in attenuating stress induced apoptosis. It is therefore suggested that ERp18 is a thiosulifide oxidoreductase involved in reducing apoptosis of stress induced cells. However, ERp18 has only been studied in in vitro and therefore, in order to gain a better understanding of the function of ERp18, in vivo studies should be conducted.
References
Alanen, H. I., et al. (2003). "Functional Characterization of ERp18, a New Endoplasmic Reticulum-located Thioredoxin Superfamily Member." J. Biol. Chem. 278(31): 28912-28920.
Jeong, W., et al. (2008). "ERp16, an endoplasmic reticulum-resident thiol-disulfide oxidoreductase: biochemical properties and role in apoptosis induced by endoplasmic reticulum stress." J. Biol. Chem.: M803804200.
Lash, A. E., et al. (2000). "SAGEmap: A public Gene Expression Resource " Genome Research 10(7): 1051-1060.
Liu, F., et al. (2003). "Isolation and characterization of a novel human thioredoxin-like gene hTLP19 encoding a secretory protein." Gene 315: 71-78.
Ye, S. Q., et al. (2002). "Microarray, SAGE and their applications to cardiovascular diseases." Cell Res 12(2): 105-115.
Bibliography
(2004). "[Mouse430_2] Affymetrix Mouse Genome 430 2.0 Array." Retrieved 2009, 14 June from http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL1261.
(2008). "Bioconductor: open source software for bioinformatics " Retrieved 14 June 2009, from http://www.bioconductor.org/overview/.
Ellgaard, L., et al. (2005). "The human protein disulphide isomerase family: substrate interactions and functional properties." Embo Reports 6(1): 28-32.
Engin, F., et al. "NOTCHing the bone: Insights into multi-functionality." Bone In Press, Accepted Manuscript.
Gao, G., et al. (2009). "Human Papillomavirus16 Variant E7 Gene Induces Transformation of NIH 3T3 Cells Via Up-Regulation of cdc25A and Cyclin A." Basic Sciences 19(4): 494-499.
Han, A., et al. (1979). "Transformation of Mouse C3H/10T1/2 Cells by Single and Fractionated Doses of X-Rays and Fission-Spectrum Neutrons." Cancer Res 39(1): 123-130.
Ishihara, H., et al. (1993). "Pancreatic beta cell line MIN6 exhibits characteristics of glucose metabolism and glucose-stimulated insulin secretion similar to those of normal islets." Diabetologia 36(11): 1139-1145.
Ishii, M., et al. (2000). "Direct Comparison of GeneChip and SAGE on the Quantitative Accuracy in Transcript Profiling Analysis." Genomics 68(2): 136-143.
Mosser, D. M., et al. (2008). "Exploring the full spectrum of macrophage activation." Nat Rev Immunol 8(12): 958-969.
Su, A. I., et al. (2004). "A gene atlas of the mouse and human protein-encoding transcriptomes." Proceedings of the National Academy of Sciences of the United States of America 101(16): 6062-6067.
Sukdeb Mondal, et al. (1976). "Two-Stage Chemical Oncogenesis in Cultures of C3H/10T1/2 Cells." Cancer Research 36: 2254-2260.
Tanabe, K., et al. (1997). "Leptin Induces Proliferation of Pancreatic [beta] Cell Line MIN6 through Activation of Mitogen-Activated Protein Kinase." Biochemical and Biophysical Research Communications 241(3): 765-768.
Walker, J. R. (2008). "C3H_10T1_2_4MJW06120828." Retrieved 14 June 2009, from http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM258729.