Evolution ERp18
This page discusses the evolution of the target protein, Endoplasmic reticulum thioredoxin superfamily member.
Introduction
importance of evolution
Methods
To generate a collection of sequences which were, apparently, related to the target protein (ERp18), a PSI-Blast search was conducted. PSI-BLAST is advantageous as it uses an iterative approach whereby selected, relavent results from previous searches are used to inform the next search operation. The PSI-BLAST method was particularly useful in this circumstance since ERp18 is part of a superfamily of proteins and so has many homologues which have high identity scores and low e-vaulues but are actually different proteins.
The collected sequences were analysed using the MEGA4 suite. This program packages allows:
- multiple sequence alignment,
- phylogenetic tree generation,
- bootstrapping, and,
- viewing and editing of phylogenetic trees
All of which were important to this assignment.
The Dayhoff Matrix was used to generate phylogenetic trees throughout this analysis
Results
Sequence Collection
The defining motif for a protein in the thioredoxin protein superfamily is the CXXC motif, with the two cystines representing the catalytic residues. It has been suggested in literature the the 'wild card' proteins between the cystines, in part, dictact the specific molecular function (see Function ERp18).
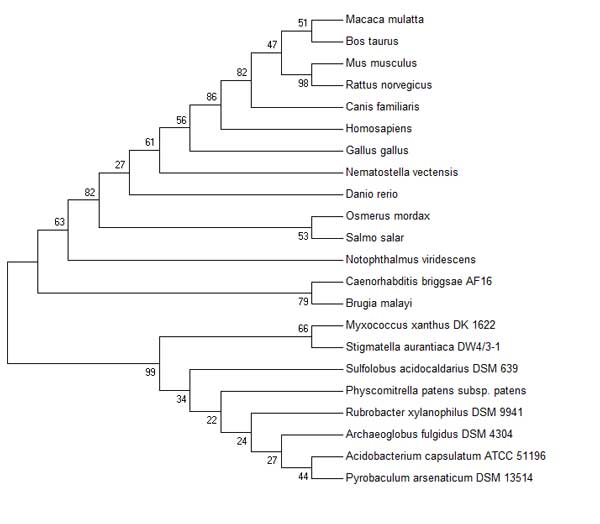
This statement is supported in part, by examining the evolution of Thioredoxin proteins in different organisms. The
[[Image:Sequence1-cml.jpg]|thumb|Evolutionary analysis of different Thioredoxin proteins showing different subfamilies]]
Phylogenetic Trees
add other zoomed in trees
Discussion
related to what organisms?
distant from other orgaisms?
important enzyme?