Pyridoxal Phosphatase Results
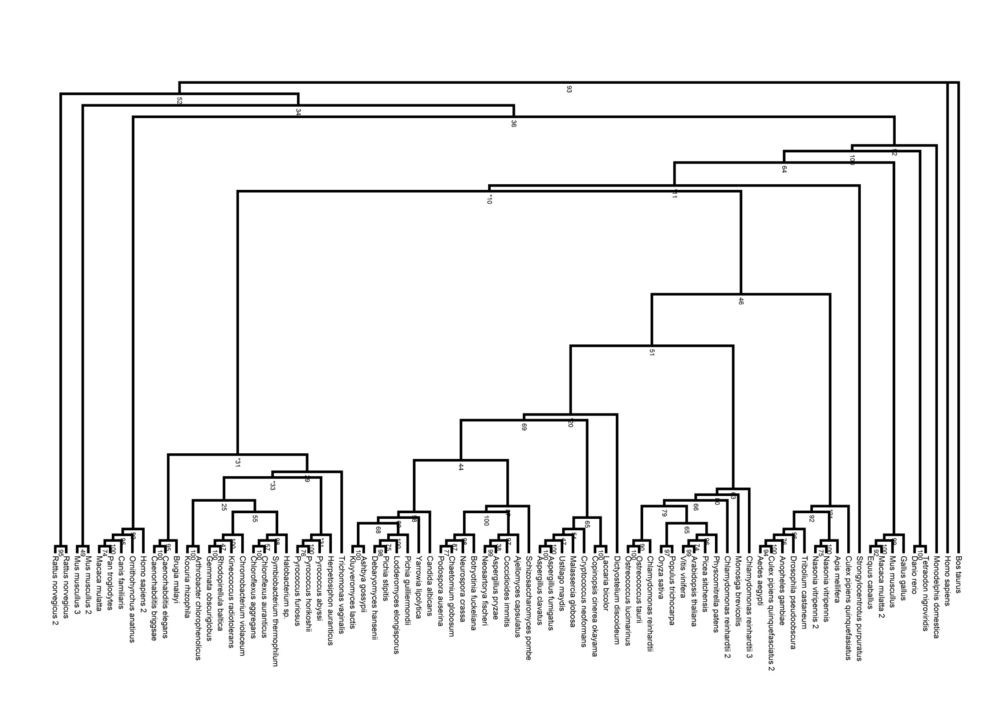
Evolution
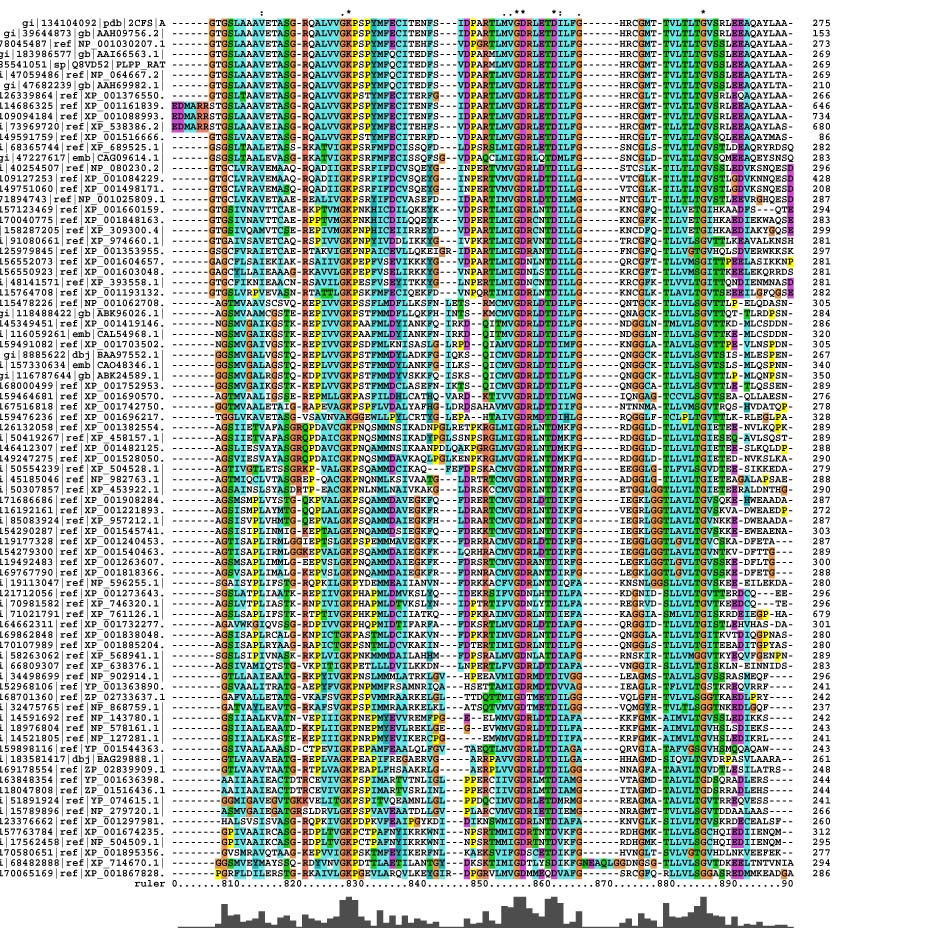
Multiple Sequence Alignment
Pyridoxal Phosphatase Multiple Sequence Alignment
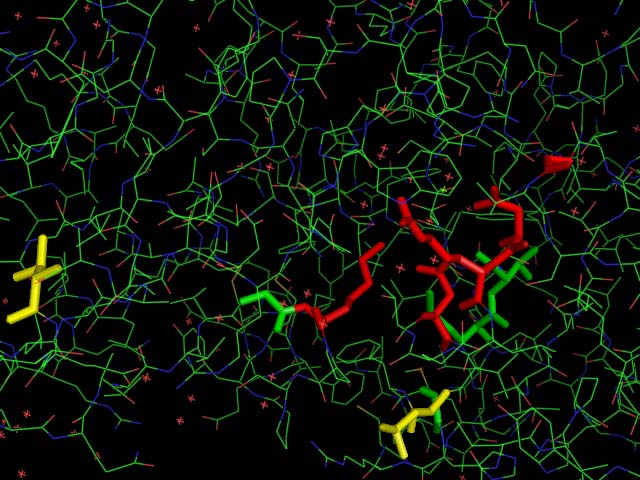
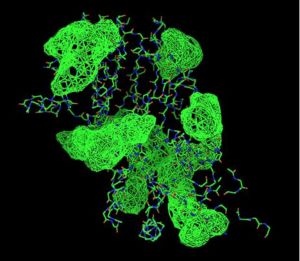
The picture on the left shows the spatial alignment of the conserved residues from the multiple sequence alignment. The red highlighted residues are those that are conserved throughout the alignment while the yellow residues are those which have conservative substitutions in some of the organisms. The green residues denote those that had semi-conservative substitutions.
Phylogeny Tree
Pyridoxal Phosphatase Phylogeny Diagram
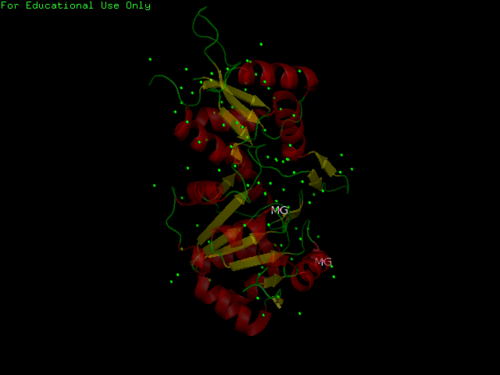
Structure
General Structure of Pyridoxal Phosphatase & 2cfsA

PDB
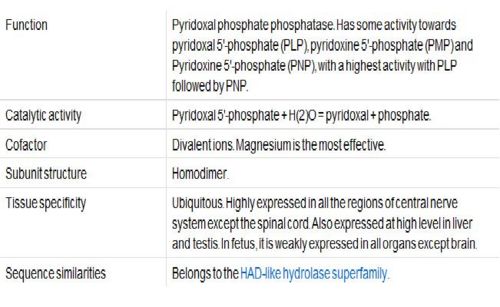
Based on the information provided in the website, Pyridoxal Phosphatase has the following characteristics:
- Pyridoxal Phosphatase is isolated from Homo Sapiens and is expressed in Escherichia Coli.
- Structure is similar to the Pyridoxal Phosphate Phospatase protein
- 1 (A) Chain
- Consists of Magnesium components
- Resolution of 2.40 angstroms. This means that the number of sidechains in the wrong rotamer is smaller as compared to proteins of a higher resolution (>2.5 angstroms). Other characteristics of proteins of similar resolutions are: (1) many small detectable errors, (2) correct folding, (3) fewer number of errors in the surface loops and (4) visible water molecules and small ligands.
http://www.rcsb.org/pdb/explore/explore.do?structureId=2cfs
DALI
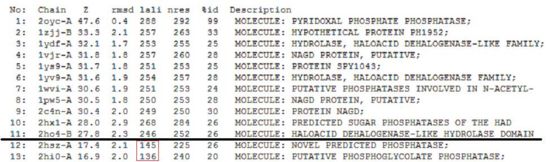
A total of 176 hits were generated, of which only the first 11 (as shown in Fig. 6) were predicted to be significant. The others were rejected on account that their respective lali scores (refer to the red, boxed section) were less than half of Pyridoxal Phosphatase's (nres: 296). The lali value is important as this refers to the number of structurally equivalent residues between the hit protein and the protein-of-interest. If two proteins are not structurally similar/related, chances are their functions will not be substantially similar.
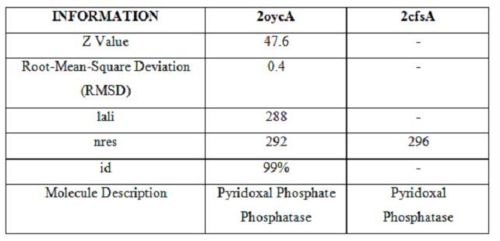
It was noted that none of the hits actually matched 2cfsA. The closest was a Pyridoxal Phosphate Phosphatase (PDB ID 2oycA) which based on the results, was predicted to be highly similar to 2cfsA. Table 1 highlights the similarities between Pyridoxal Phosphatase (2cfsA) and the Pyridoxal Phosphate Phosphatase (2oycA)
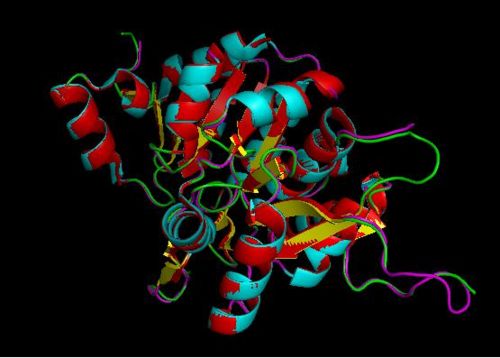
Using the PyMOL software, 2oycA was superimposed against 2cfsA, and both structures, as shown in Fig. 7, are structurally similar.
Towards the end of the study, 2cfsA was re-run against the DALI database. The rationale behind doing so was that the initial results yielded 2oyc as the query protein.
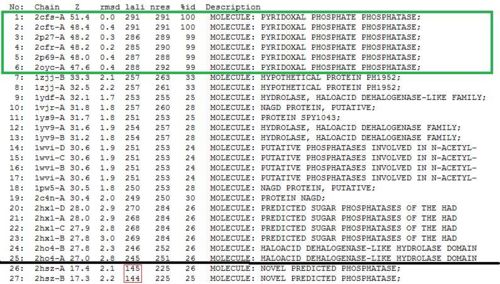
As opposed to the first run, 515 hits were generated. The top 25 protein hits were deemed significant, with the 26th protein hit onwards rejected for the same reason as that of the first batch - that their lali values were too low (less than half of 2cfs') for them to be considered as structurally significant to 2cfsA. More importantly was the observation that the DALI database identified 2cfsA as the query protein (unlike the first run). In addition, protein hit numbers 2 (2cftA) - 6 (2oycA) were ALL observed to belong to the Pyridoxal Phosphate Phosphatase family. Based on this, it was easier to narrow down the potential research candidates in terms of structural similarities. Due to time constraint however, not all the protein hits were researched on.
PDBsum
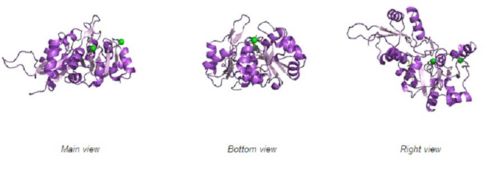
Fig. 8 offers three different views of 2cfsA. The purple chains represent the amino acid chain of 293 amino acids while the green spheres represent the magnesium ions (x2).
By clicking the "Protein chain" link, the user was re-directed to a website containing information pertaining to the secondary structures of both 2cfsA and 2oycA.
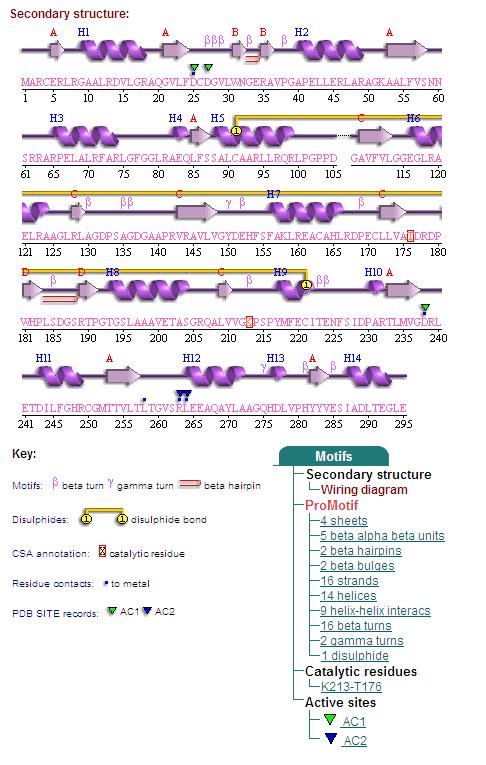
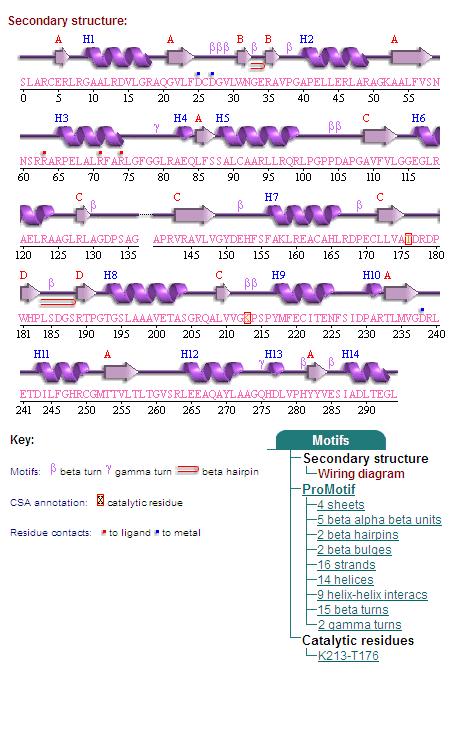
Secondary Structures of 2cfsA (L, http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl) and 2oycA (R).
Topology diagrams of 2cfsA (Fig 9.1. http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=2cfs&template=protein.html&r=wiring&l=1&chain=A) and 2oycA (Fig 9.2). The topology diagram is a simplified version of the secondary structure information as provided above, and is an indication of the location of the alpha helices (represented by the red cylinders) and beta strands (represented by the pink arrows).
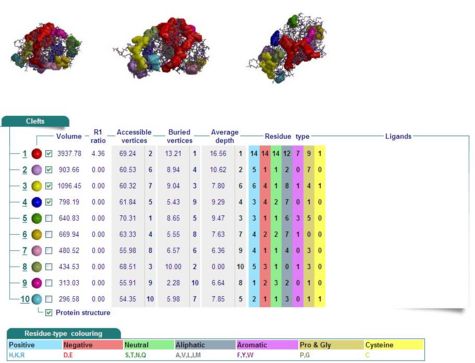
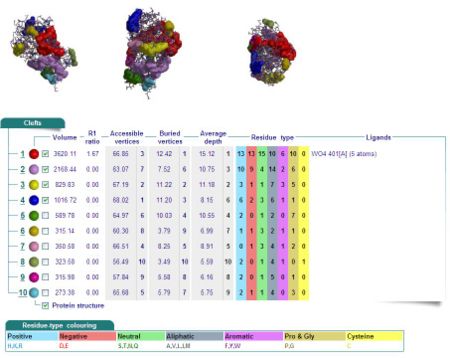
Cleft Analyses of 2cfsA (Fig 10.1. http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=2cfs&template=clefts.html&pdbcode=2cfs&r=speedfill) and 2oycA (Fig 10.2).
Cleft Analysis via PyMol
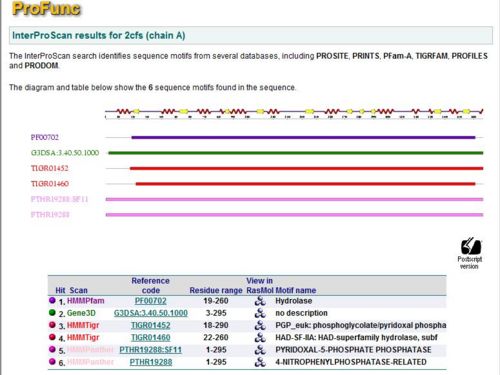
PROFUNC (2cfsA)
Related Protein Sequences in the PDB (SAS)

Matches to existing PDB Structures
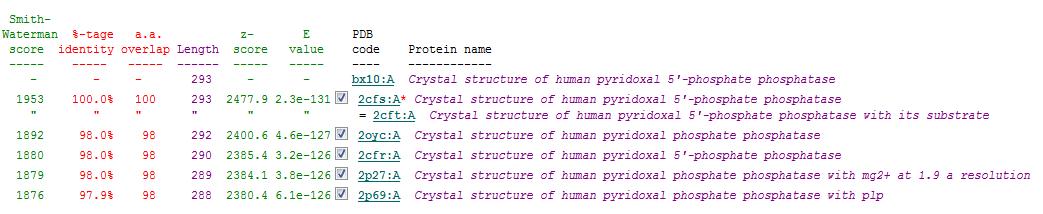
Secondary Structure Matching (SSM)
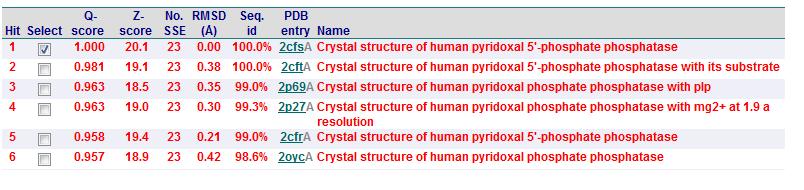
Based on the results obtained via the local alignment method (Smith-Waterman method), it was noted that 2fcsA and 2oycA were indeed similar in terms of sequence and structure. However, contrary to the implications of the DALI-generated results, 2oycA was no longer the most closely-related protein to 2cfsA. That role now belonged to 2cftA. Based on the results generated by Profunc, 2cftA's sequence and structure, as provided by PDB, were identical to that of 2cfsA. The secondary structure matching, however, showed that 2cfsA and 2cftA had differing (albeit minimally) Q scores (1.000 and 0.981 respectively). 2cfsA was described as a "Crystal structure of human pyridoxal 5'-phosphate phosphatase", while 2cftA was described as a "Crystal structure of human pyridoxal 5'-phosphate phosphatase with its substrate". In addition, 2oycA was rated 4 tiers below 2cftA (second in the scoring table).
PROFUNC (2oycA)
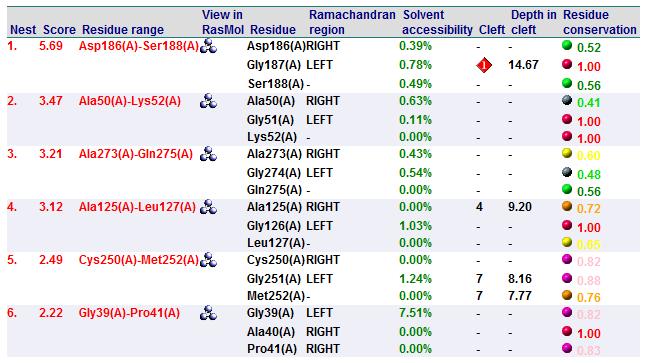
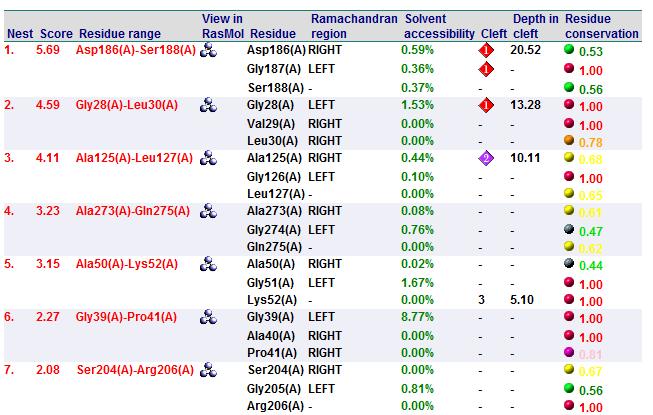
A search on 2oycA was also carried out via PROFUNC, and consistency in the results confirmed that 2cfsA and 2oycA were indeed structurally similar. This lends weight to the original hypothesis that they could be functionally related. In fact, based on the Nest Analysis method, 2oycA was observed to be sharing a large number of active sites. The results of the Nest Analysis (2oycA) are as shown below. Notice the similarities between the active sites of both 2oycA and 2cfsA.
In addition, the predicted function of 2oycA was similar to 2cfsA.
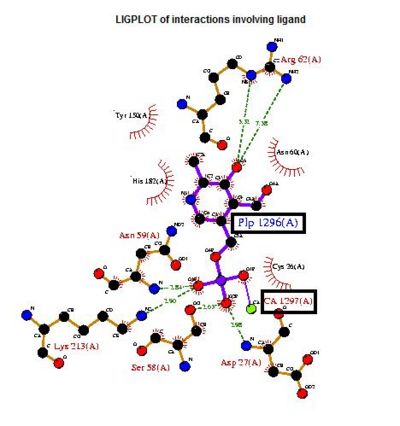
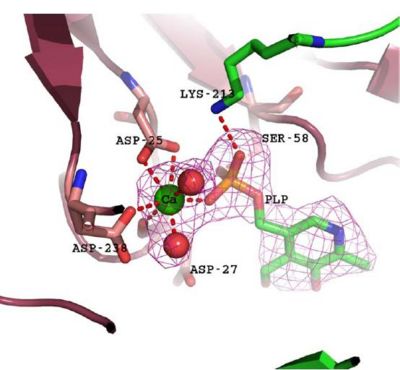
Based on the above-mentioned results, it was concluded that 2cfsA, 2cftA and 2oycA were structurally similar, and that further functional studies could indeed uncover the function of the protein-of-interest: 2cfsA
Function
In order to predict the function of Pyrixol Phosphotase, sequence homology, structural similarities, neighbouring genes, expression levels in tissue and protein-protein interactions were analysed.
General Annotation
Sequence homology
To check for function among similar homologs, 2cfs_A sequence was run against FASTA, Pfam and PSI Blast non-redundant database. A search using Pfam and BLAST revealed that 2cfs_A belonged to the superfamily, Haloacid dehalogenase-like hydrolase. The top 15 search results from BLAST
http://blast.ncbi.nlm.nih.gov/Blast.cgi
Structural Similarities
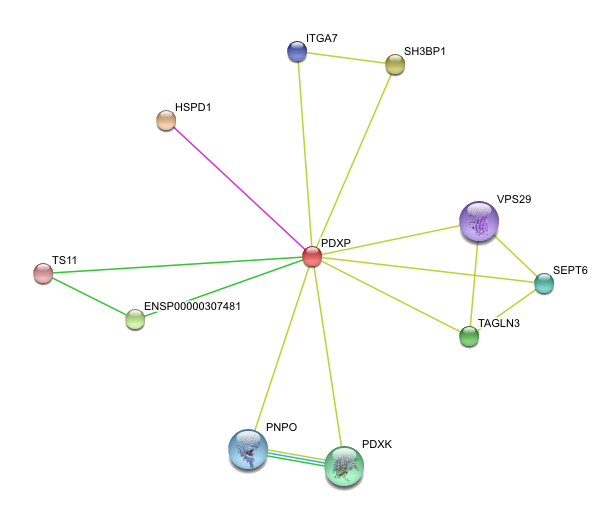
Protein-Protein Interactions
http://string.embl.de/newstring_cgi/show_network_section.pl?taskId=ph89gQxcLucL&allnodes=1