2gnx Discussion
Evolutionary Analysis
From the results it can be seen that all instances of this protein are found in metazoans and mostly in higher metazoans. The protein is also found in some insects however they are distantly related to the target sequence (2GNX A) used as can be seen from the phylogenetic tree (Figure 2). It is also interesting that this protein was not found in drosophila or C. elegance as both of these organisms have had there genome fully sequenced. The fact that this protein is mostly represented but higher metazoans indicates that this is a relatively new protein evolutionary and one that is not crucial to survival. This protein however appears to be very highly conserved amongst the higher metazoans possibly suggesting that there is selection pressure on it to remain unchanged.
Structural Analysis
The structural analysis of the 2GNX protein did not produce any significant matches upon submission to the Dali database. Therefore, a separate analysis of the N-terminal domain and C-terminal domain of the protein was carried out. Results from these analyses showed us that there were no significant matches to the N-terminal domain of the protein. Much research and analysis into the closest search results generated by the Dali database was carried out using various resources. However, these structural matches (such as the structural match with 2CMR:A protein) gave little infomation regarding to the protein function. It was then concluded that much more thorough research and analysis is required in order to identify the structure of the N-terminal domain of the protein.
An analysis of the C-terminal domain of the protein using the Dali database was able to provide us with some significant structural matches, of high interest being the GAF signalling protein. A CE analysis confirmed that the structure of the GAF protein did indeed match closely to the C-terminal domain of the 2GNX protein. Since a relation to olfactory transduction had been found through functional analysis, particular emphasis was put into investigating the liklihood of cAMP or cGMP binding. Dotlet analysis was indicated no internally homogous repeats in 2GNX, which meant 2GNX was not similar to any typical cGMP-binding phosphodiesterases (Ho YS et al, 2000).
It identified that the GAF protein contained a special cGMP ligand binding pocket (Martinez et al, 2002). Using the computational biology software PyMol it was identified that while the 2GNX protein did not contain an exact match of the ligand binding pocket (Figure 8 in the result section), there were a few residues that did match (Zoraghi R, 2003). Therefore it was likely that the protein does bind with a cGMP ligand. However, since twenty-seven residues that might comprise the ligand binding pocket of the protein were not located in its PDB file, further research and analysis has to be carried out to confirm that the protein does indeed contain a cGMP ligand-binding pocket.
Functional Analysis
Despite most of the conventional tools failing to return significant results, multiple resources suggest that this protein is related or a part of the olfactory transduction pathway. The results from the symatlas were the first hint of this possibility. Following this, the cis-RED motif results showed that this hypothesis was quite significant.
cis-RED is a database which holds conserved sequence motifs. Motifs, sometimes referred to as modules, are conserved cis-regulatory regions which contain the spatially clustered binding sites of transcription factors. The cis-RED analysis presented 14 modules present in the BC048403 protein. All of these modules are also found in olfactory receptors co-occurring in groups from one to nine. Up to 120 olfactory receptors corresponded to these 14 modules and the most significant results - those with 3 or more co-occurring modules, appear in Table 6 of the results. The two most significant olfactory receptors had 9 modules in common with the BC048403 protein. The top five co-occuring modules were investigated in terms of conservation and expression, however no further results were gained.
Further evidence of this proteins involvement in olfactory transduction is the Micro-Array profiles shown in Figure 6 of the results. These expression profiles were found by searching the neighbouring profiles of the human ortholog FLJ32549. We were hopeful of finding a corresponding olfactory receptor to those found through the cis-RED analysis, however this did not occur.
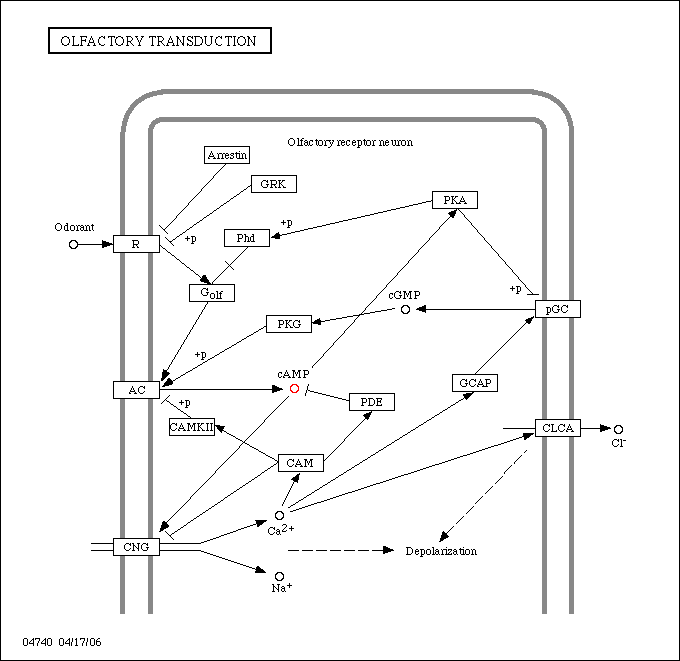
The following image represents the olfactory transduction pathway. Olfactory transduction is initiated by a membrane protein and contains a typical cAMP cycle. cGMP is also a mediator of the olfactory transduction pathway.
From the structural analysis, since it was found that cGMP binding is possible, the cis-regulatory elements were checked for involvement in the olfactory transduction pathway. Several modules were found to be related to genes that could well be involved in the olfactory transduction pathway. Within several of the modules a type I cGMP-dependent protein kinase was found.
It is our hypothesis that this protein is involved in the olfactory transduction pathway and in particular, being inolved in the cGMP cycle. Due to the expression data and locate analysis being varied it is not possible to hypothesize as to what exact part of an organism the protein functions in.
Return to Report