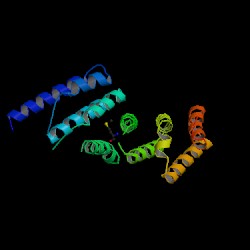
2i2O Protein Structure Presentation
Materials and Methods
1) Protein search on PDB, NCBI Entrez, Pfam and InterPro databases
- PDB code used: 2i2o
2) Identifying other similar structures with DALI server
3) Comparing the identified structures using CE comparison
Results
PDB
- Protein was isolated form Danio rerio and expressed in Escherichia coli.
- Structure has similarities with the “Eukaryotic Translation Initiation Factor 4G (eIF4G)” protein.
- Protein has 2 chains; and consists of nickel (Ni2+) and selenomethionine (C5H11NO2Se) components.
Pfam and InterPro
- Identified as "Middle domain of eIF4G", termed MIF4G.
- Consists essentially of alpha helices and has “multiple alpha-helical repeats”. Within eIF4G, it binds to the RNA helicase eIF4A, eIF3, RNA and DNA.
DALI results
- SUMMARY: PDB/chain identifiers and structural alignment statistics
NR. STRID1 STRID2 Z RMSD LALI LSEQ2 %IDE REVERS PERMUT NFRAG TOPO PROTEIN 1: 3028-A 2i2o-A 37.1 0.0 206 206 100 0 0 1 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION eif4g-like prote 2: 3028-A 1hu3-A 15.2 2.7 164 204 23 0 0 12 S TRANSLATION eif4gii fragment (eukaryotic initiation f 3: 3028-A 1uw4-B 10.9 3.6 164 247 9 0 0 12 S NONSENSE MEDIATED MRNA DECAY PROTEIN regulator of nons 4: 3028-A 1h6k-A 10.6 3.9 175 728 11 0 0 11 S NUCLEAR PROTEIN cbp80 fragment (ncbp 80 kda subunit, 5: 3028-A 2db0-A 8.1 3.3 148 239 14 0 0 13 S PROTEIN BINDING 253aa long hypothetical protein (hypot 6: 3028-A 1b3u-A 8.0 27.7 150 588 13 0 0 13 S SCAFFOLD PROTEIN protein phosphatase pp2a fragment
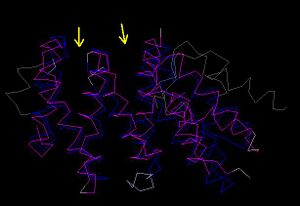
CE Structural comparison
- 1hu3 and 2i2o: Alpha-helices between 2i2o and 1hu3 were folded in same orientation
- 1h6k and 2i2o: N-terminal region of 1h6k displayed similarity with 2i2o; hence a possibility that MIF4G protein is a region, or even an active domain, near the N-terminal of the nuclear cap binding protein complex (CBC).
Discussion
- Protein of interest is highly similar in structure to human MIF4G (PDB: 1hu3). Therefore, MIF4G can be utilised as a model to formulate predictions on its structure.
- The MIF4G contains five helical hairpins oriented in a right-handed solenoid, and is similar to the HEAT [Huntingtin, elongation factor 3, a subunit of protein phosphatase 2A (PP2A), and target of rapamycin] domain (Marintchev & Wagner 2005).
- Protein has an overall crescent shape. Helical hairpins are stacked on top of the other to confer this shape.
- MIF4G binds to eIF4A, an RNA helicase, and RNA, hence rendering its role in regulating cell translation. A protease-resistant region identified by proteolysis and mass spectrometry is speculated to be the binding site for eIF4A (Marcotrigiano et al, 2001).
- MIF4G is one of the 3 domains on eIF4G RNA regulatory protein. Each of these domains are connected by linkers.
- eIF4G has 3 domains: MIF4G, MA3 and W2
- It was hypothesized that the C’ terminal linker segment of MIF4G interacts with the domain by wrapping around the inter-helical grooves of the domain (Marintchev & Wagner 2005). This will account to an extent the presence and potential function of the numerous grooves observed in the domain structure.
- All further hypotheses on structural organization and interactions of the domain within the eIF4G protein necessitate additional analysis.