ATP binding domain 4 Discussions: Difference between revisions
Anharmustafa (talk | contribs) |
|||
| (8 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
'''Multiple sequence alignment''' | '''Multiple sequence alignment''' | ||
From the multiple sequence alignment, there are seven conserved regions found in all species from Domain Eukarya and Archaea (based on | From the multiple sequence alignment, there are seven conserved regions found in all species from Domain Eukarya and Archaea (based on BlastP search) but only five conserved residues which are significant to the function and structure of the protein since these residues are identified to be conserved amino acid sequence motif for P-loop of nucleotide binding domains which might be important in phosphate binding (Bork and Koonin, 1994). These conserved residues are Serine-12, Glysine-13 and Glysine-14, Lysine-15 and Aspartic acid-16 . Since this motif presence in uncharacterized ATP pyrophosphatase domain, the motif is called PP motif and can be written as S-G(2)-K-D-[GS]. PP-loop motif is a modified version of the P-loop of nucleotide binding domain that is involved in phosphate binding (Bork and Koonin,1994). However, based on the multiple sequence alignment, the amino acids for PP-loop that are conserved across all species in Eukarya and Archaea are Glysine-12 and Glysine-14. Substitution of Serine with Threonine in ''Pyrobaculum arsenaticum, Pyrobaculum calidifontis, Thermoproteus neutrophilis, Pyrobaculum islandicum'' and ''Staphlothermus marinus'', are conserved since both amino acids are polar uncharged amino acid, suggesting that this residue is also important. However, this residue is not conserved in ATP Binding Domain 4 in ''Homo sapiens'' which is only encoded by 259 amino acid sequences and the sequence started from Glysine-13. Moreover, amino acid in residue 17 is also one of the important residues in PP motif. This residue is conserved across all species except for ''Theileria parva'' and ''Theileria annulata'' where Serine is substituted with Glysine. | ||
BlastP search revealed the very low E value from many sequences from different species. These sequences appear to be encoding different group of proteins such as ATP pyrophosphatase, ATP-binding protein, ATPase and endoribonuclease. Although these proteins are responsible for different functions, the similarities of sequence between the proteins across species are very high (as indicated by E value) and the sequences which are conserved are a part of PP motif, suggesting that these proteins descend from a common ancestral sequence and therefore are paralogs. Such phenomenon could be as the result of duplication of genes within a genome. | |||
''' | '''Phylogenetic tree and Bootstrap''' | ||
The | The phylogenetic tree and bootstrap revealed that PP-loop motif from ATP binding domain 4 (belongs to N-type ATP pyrophosphatase/ATPases) and other related proteins are found in species of Archaea and Eukarya suggesting that this motif is highly conserved throughout evolution. However, Bacteria found to be lacking of this conserved sequences since none of the species belongs to Domain Bacteria. Nevertheless, based on STRING: functional protein association networks, some bacteria species still have protein sequences which belong to N-type ATP pyrophosphatase superfamily. BlastP was conducted to compare the protein sequences of ATPases from ''Pyrococcus furiosus'' and the bacteria species which under the clan of N-type ATP pyrophosphatase. The E value obtained from the result are below the E-value that is used as the cut-off-score. The bacteria species are ''Fusobacteruim nucleatum'' (1e-22), ''Caldicellulosiruptor saccharolyticus'' (1e-13),''Campylobacter jejuni'' (4e-19),''Chromobacterium violaceum'' (1e-21)and ''Polynucleobacter sp.'' (1e-24). These E value fall below the lowest E value based on BlastP from ''Pyrococcus furiosus'' and ''Macacca mulatta'' i.e: 5e-50 and 2e-25 respectively. These bacteria species can not be found in BlastP on the first place because the E value are quite high which indicate the sequence similarity between bacteria species and ''Pyrococcus furiosus'' and ''Macaca mulatta'' are very low. These suggested that although some bacteria species still have protein sequences which belong to N-type ATP pyrophosphtase superfamily, they are distantly related to both archaea and eukarya. Based on the high protein sequence similarity between Archaea and Eukarya, it can be suggested that Archaea and Eukarya are closely related compared to that for Bacteria. | ||
Based on unrooted tree, the relatedness of the taxa and their relationship can be illustrated but the last common ancestor and how the species evolved cannot be observed. | Based on unrooted tree, the relatedness of the taxa and their relationship can be illustrated but the last common ancestor and how the species evolved cannot be observed. It was found that the taxa are clearly grouped into Domain Archaea and Eukarya. Moreover, in Eukarya, the taxa are nicely clustered according to parasites, vertebrates and invertebrates. Domain Bacteria is missing in this tree suggesting that Bacteria are distantly related to Archaea and Eukarya based on the sequence of ATP Binding Domain 4. | ||
Phylogram is another way to illustrate the evolution of the taxa where the relationships between the taxa and | Phylogram (figure 3.0) is another way to illustrate the evolution of the taxa where the relationships between the taxa and the time or rate of evolution can be observed. | ||
Bootstrap was conducted to test the reliability of the branching order of the | Bootstrap was conducted to test the reliability of the branching order of the phylogenetic tree. Based on the bootstrap value, we can be confident with the order of the phylogenetic tree thus allowing us to determine where speciation events occur. Bootstrap worked by performing 'pseudoreplicates' of multiple sequence alignments and in this project, 100 replicates was performed. The bootstrap tree can then be generated thus comparing the branching orders and distances of phylogenetic tree. The bootstrap value for each branch are obtained in percentage which indicate the confidence of the branch being correct. If the value of bootstrap is less than 75%, the branching order is not very reliable and meaningless. If the value is between 90% and above, we can be confident that the branching orders are correct. Based on the bootstrap result, it was found that most of the value are lower than 75% suggesting that the branching patterns and distances are not reliable. Therefore, another phylogenetic tree need to be built in order to increase the reliability of the branching pattern and distances of the tree. | ||
Based on unrooted tree, it was found members of | Based on unrooted tree, it was found members of Domain Eukarya are nicely cluster at one side without any presence of other species from different domain i.e: members from different domains are well-seperated. Therefore, it can be suggested that the evolutionary model for the PP-loop motif is hold since there is no evidence of the occurrence of lateral gene transfer. | ||
These result support theory for origins of eukaryotes which suggested chimeric features of eukaryote genome (fusion of archaea and bacteria). Study which involved whole-genome-sequence data to test this theory, indicates Eukaryote genome is a chimera of genes most similar to that in Archaea and Bacteria. This study used 'homology-hit' analysis in which the genes from eukaryotes from different classes matching to nearest homology genes in Archaea and Bacteria. It was found that informational genes are closely related to Archaea whereas operational genes are closely related to bacteria (Horiike et al., 2001). Moreover, study conducted by Brown and Doolittle which involved analysing geneolgies from 66 protein-coding genes from members of all three domains of life found that Arginosuccinate synthase from Eukaryotes are closely related to Archaea (analysis on structural similarity found that the conserved region of Arginosuccinate synthase are similar to that of ATP pyrophosphatase)(Katz, 1998). Hence, finding from previous studies support the evolutionary relationship between the three Domain of life based on ATP Binding Domain 4 which suggested that that Archaea and Eukarya are closely related. | |||
== '''Discussion''' == | [[Image:Horiike.PNG|left|thumb|1200px|'''Figure 6.0''': Chimeric nature of eukarya based on geneologies. Figure adapted from Katz, 1998 ]] | ||
<BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR> | |||
== '''Discussion On Structure''' == | |||
ATP binding Domain 4 (IRU8) is a protein with an unknown function. The crystallization of 1RU8 was done from Pyrococcus furiosus that is expressed in Escherichia Coli with resolution of 2.7 Angstroms and an R-value of 0.218. Artificial chemical ligand, TRS(2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL) was used in the crystallization in order to maintain the integrity of the structure when crystallized. There is no actual ligand of this protein based on the the literature paper and from the Protein Data Base (PDB). This is probably because the function of the protein is still unknown. Determination of the ligand can also leads to the possible function of the protein. Hence, deduction of the ligand of this protein can only be done, based on other protein's ligand that has similar function or structure. | ATP binding Domain 4 (IRU8) is a protein with an unknown function. The crystallization of 1RU8 was done from Pyrococcus furiosus that is expressed in Escherichia Coli with resolution of 2.7 Angstroms and an R-value of 0.218. Artificial chemical ligand, TRS(2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL) was used in the crystallization in order to maintain the integrity of the structure when crystallized. There is no actual ligand of this protein based on the the literature paper and from the Protein Data Base (PDB). This is probably because the function of the protein is still unknown. Determination of the ligand can also leads to the possible function of the protein. Hence, deduction of the ligand of this protein can only be done, based on other protein's ligand that has similar function or structure. | ||
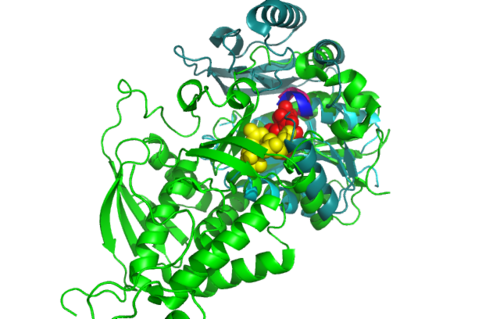
[[Image:Front view.png|left|thumb|500px|'''Figure 5.0'''. Structure alignment between 1RU8 (magenta) and 2NZ2 (green) generated via PyMol-Front view. Highlighted yellow and red is the Citrulline and the ATP molecule respectively. Meanwhile the highlighted colour of pink and blue represent the conserved region of PP-loop.]] | |||
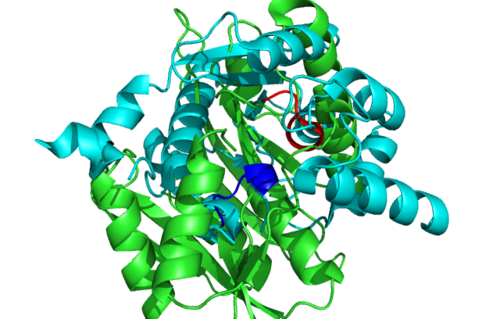
[[Image:1RU8 and 3BL5.png|right|thumb|500px|'''Figure 6.0'''. Structure alignment between 1RU8 (magenta) and 3BL5 (green) generated via PyMol. Highlighted colour of blue and red represent the conserved region of PP-loop.]] | |||
<BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR> | |||
From the structure obtained from PyMOL, visualization of the secondary structure provide information on the surface properties covering positively charged regions and negatively charged regions (figure 3.0), its domains (figure 2.0) as well as ligand binding sites and surface clefts (figure 7.0), and also conservation of residues across different species(figure 2.0). It was observed that detailed secondary structure can be determined using PDBsum where conserved residues, protein motif and possible ligand binding site was highlighted (Figure 2.0). Information from the PDB and SCOP has indicated that 1RU8 is from the family of N-type ATP pyrophosphatases and also under the clan of PP-loop which is the strongly conserved motif Ser(12),Gly(13),Gly(14),Lys(15) and Asp(16) at the N terminus (Bork & Koonin 1994). This PP-loop was significant as it is the most conserved and is involved in ligand bindings and thus is likely to give function to our protein. Besides that, from CATH domain database, two main domains were found in 1RU8. The first domain ranges from residue 3-97 while the second domain residues from 98-232. Topology of Domain 1 is known to be of Rossmann A-B-A fold, and the superfamily of PP-loop is presence in this domain. Domain 2 is A-B complex classified in CATH. | From the structure obtained from PyMOL, visualization of the secondary structure provide information on the surface properties covering positively charged regions and negatively charged regions (figure 3.0), its domains (figure 2.0) as well as ligand binding sites and surface clefts (figure 7.0), and also conservation of residues across different species(figure 2.0). It was observed that detailed secondary structure can be determined using PDBsum where conserved residues, protein motif and possible ligand binding site was highlighted (Figure 2.0). Information from the PDB and SCOP has indicated that 1RU8 is from the family of N-type ATP pyrophosphatases and also under the clan of PP-loop which is the strongly conserved motif Ser(12),Gly(13),Gly(14),Lys(15) and Asp(16) at the N terminus (Bork & Koonin 1994). This PP-loop was significant as it is the most conserved and is involved in ligand bindings and thus is likely to give function to our protein. Besides that, from CATH domain database, two main domains were found in 1RU8. The first domain ranges from residue 3-97 while the second domain residues from 98-232. Topology of Domain 1 is known to be of Rossmann A-B-A fold, and the superfamily of PP-loop is presence in this domain. Domain 2 is A-B complex classified in CATH. | ||
| Line 40: | Line 50: | ||
[[Image:AS.gif |left|thumb|800px|'''Figure 8.0'''. Argininosuccinate synthetase (AS)catalyic mechanism. ]]<BR><BR><BR><BR><BR><BR><BR> | [[Image:AS.gif |left|thumb|800px|'''Figure 8.0'''. Argininosuccinate synthetase (AS)catalyic mechanism. ]]<BR><BR><BR><BR><BR><BR><BR><BR><BR> | ||
<BR><BR> | |||
From the figure 8.0, | From the figure 8.0, | ||
| Line 56: | Line 65: | ||
The structure of AS is used for the modelling ATP binding into our structure. The beta and gamma phosphate groups of ATP are oriented by characteristic residues of the PP-loop, and then forming a salt linkage between the g-phosphate and the N atom of lysine. (Lemke & Howell 2001) | The structure of AS is used for the modelling ATP binding into our structure. The beta and gamma phosphate groups of ATP are oriented by characteristic residues of the PP-loop, and then forming a salt linkage between the g-phosphate and the N atom of lysine. (Lemke & Howell 2001) | ||
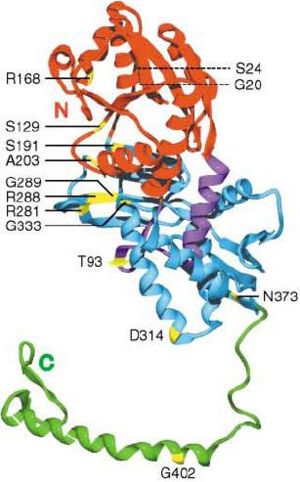
[[Image:AS3.JPG |left|thumb| | [[Image:AS3.JPG |left|thumb|300px|'''Figure 8.1'''. Proposed important residues for ATP or substrate binding in argininosuccinate synthetase.]] | ||
[[Image:1RU8 conserved residues1 huhu.PNG|centre|thumb|700px|'''Figure 8.2'''. Conserved residues of ATP Binding Domain 4 generated via PyMol that may have similar role to AS.]] | |||
<BR><BR><BR> | |||
In the AS model in E. coli., R168 (see figure 8.1) has been proposed for pyrophosphate binding. The carbonyl oxygen of the second residue preceding the PP motif in AS (Ala-16), falls within the hydrogen bonding distance of the O2’ hydroxyl oxygen of the ribose. (Lemke & Howell 2001). Therefore, the alanine residue closely after the PP-motif in our protein, Ala(20) should be involved in stabilising ribose by forming H-bond with its O2’ hydroxyl oxygen. This may explain why relatively majority of aliphatic residues are conserved in the biggest gap in our cleft analysis (see figure 4.2 under function analysis) and relatively highly conserved detected by PDBsum. | In the AS model in E. coli., R168 (see figure 8.1) has been proposed for pyrophosphate binding. The carbonyl oxygen of the second residue preceding the PP motif in AS (Ala-16), falls within the hydrogen bonding distance of the O2’ hydroxyl oxygen of the ribose. (Lemke & Howell 2001). Therefore, the alanine residue closely after the PP-motif in our protein, Ala(20) should be involved in stabilising ribose by forming H-bond with its O2’ hydroxyl oxygen. This may explain why relatively majority of aliphatic residues are conserved in the biggest gap in our cleft analysis (see figure 4.2 under function analysis) and relatively highly conserved detected by PDBsum. | ||
| Line 64: | Line 76: | ||
The most intriguing part is in the highly conserved PP-loop motif ([A/S]-[F/Y]-S-G-G-[L/V]-D-T-[S/T]) contains two absolutely conserved glycine residues, Gly(13) and Gly(14). In AS model, the two conserved glycine residues showed significantly different conformations in its uncomplexed and complexed structures, which are suspected to play a role in binding and release of pyrophosphate (PPi). (Lemke & Howell 2001) This agrees with glycine, lacking side chains, can provide a high degree of flexibility. Therefore, Gly(13)Gly(14) in ATP binding domain 4 should provide a large anion hole required for pyrophosphate binding. Lemke and Howell (2001) proposed that other N-type ATP pyrophosphatases models, if having glycine residues replaced, the steric hindrance created will crash with the bridge oxygen of the bound pyrophosphate. | The most intriguing part is in the highly conserved PP-loop motif ([A/S]-[F/Y]-S-G-G-[L/V]-D-T-[S/T]) contains two absolutely conserved glycine residues, Gly(13) and Gly(14). In AS model, the two conserved glycine residues showed significantly different conformations in its uncomplexed and complexed structures, which are suspected to play a role in binding and release of pyrophosphate (PPi). (Lemke & Howell 2001) This agrees with glycine, lacking side chains, can provide a high degree of flexibility. Therefore, Gly(13)Gly(14) in ATP binding domain 4 should provide a large anion hole required for pyrophosphate binding. Lemke and Howell (2001) proposed that other N-type ATP pyrophosphatases models, if having glycine residues replaced, the steric hindrance created will crash with the bridge oxygen of the bound pyrophosphate. | ||
The final residue of the PP motif in our structure, the hydroxyl group of Ser(17) should be involved in forming hydrogen bonds with the highly conserved residues, Gly(14) and Arg(60), meaning Ser(17) residue is important both for pyrophosphate binding and structural support by interacting with a helices, therefore H1 and H2 can be linked together. (see Figure 2.0 showing H1 and H2 in PDBsum) | The final residue of the PP motif in our structure, the hydroxyl group of Ser(17) should be involved in forming hydrogen bonds with the highly conserved residues, Gly(14) and Arg(60), meaning Ser(17) residue is important both for pyrophosphate binding and structural support by interacting with a helices, therefore H1 and H2 can be linked together. (see Figure 2.0 showing H1 and H2 in PDBsum) | ||
<BR><BR> | |||
(2) '''Protein conformational change existed in the catalytic cycle.''' | (2) '''Protein conformational change existed in the catalytic cycle.''' | ||
| Line 90: | Line 102: | ||
[[ATP binding domain 4 Functions | Functional Analysis]]| | [[ATP binding domain 4 Functions | Functional Analysis]]| | ||
[[ATP binding domain 4 Evolution | Evolutionary Analysis]]|<BR> | [[ATP binding domain 4 Evolution | Evolutionary Analysis]]|<BR> | ||
[[ATP binding domain 4 Discussions | Discussions]]| | [[ATP binding domain 4 Discussions | Discussions]]| | ||
[[ATP binding domain 4 Conclusion | Conclusion]] | | |||
[[ATP binding domain 4 References | References]] | [[ATP binding domain 4 References | References]] | ||
[[ATP binding domain 4 | Back to Main ATP binding domain 4 pages]] | [[ATP binding domain 4 | Back to Main ATP binding domain 4 pages]] | ||
Latest revision as of 09:05, 14 June 2009
Discussion on Evolution
Multiple sequence alignment
From the multiple sequence alignment, there are seven conserved regions found in all species from Domain Eukarya and Archaea (based on BlastP search) but only five conserved residues which are significant to the function and structure of the protein since these residues are identified to be conserved amino acid sequence motif for P-loop of nucleotide binding domains which might be important in phosphate binding (Bork and Koonin, 1994). These conserved residues are Serine-12, Glysine-13 and Glysine-14, Lysine-15 and Aspartic acid-16 . Since this motif presence in uncharacterized ATP pyrophosphatase domain, the motif is called PP motif and can be written as S-G(2)-K-D-[GS]. PP-loop motif is a modified version of the P-loop of nucleotide binding domain that is involved in phosphate binding (Bork and Koonin,1994). However, based on the multiple sequence alignment, the amino acids for PP-loop that are conserved across all species in Eukarya and Archaea are Glysine-12 and Glysine-14. Substitution of Serine with Threonine in Pyrobaculum arsenaticum, Pyrobaculum calidifontis, Thermoproteus neutrophilis, Pyrobaculum islandicum and Staphlothermus marinus, are conserved since both amino acids are polar uncharged amino acid, suggesting that this residue is also important. However, this residue is not conserved in ATP Binding Domain 4 in Homo sapiens which is only encoded by 259 amino acid sequences and the sequence started from Glysine-13. Moreover, amino acid in residue 17 is also one of the important residues in PP motif. This residue is conserved across all species except for Theileria parva and Theileria annulata where Serine is substituted with Glysine.
BlastP search revealed the very low E value from many sequences from different species. These sequences appear to be encoding different group of proteins such as ATP pyrophosphatase, ATP-binding protein, ATPase and endoribonuclease. Although these proteins are responsible for different functions, the similarities of sequence between the proteins across species are very high (as indicated by E value) and the sequences which are conserved are a part of PP motif, suggesting that these proteins descend from a common ancestral sequence and therefore are paralogs. Such phenomenon could be as the result of duplication of genes within a genome.
Phylogenetic tree and Bootstrap
The phylogenetic tree and bootstrap revealed that PP-loop motif from ATP binding domain 4 (belongs to N-type ATP pyrophosphatase/ATPases) and other related proteins are found in species of Archaea and Eukarya suggesting that this motif is highly conserved throughout evolution. However, Bacteria found to be lacking of this conserved sequences since none of the species belongs to Domain Bacteria. Nevertheless, based on STRING: functional protein association networks, some bacteria species still have protein sequences which belong to N-type ATP pyrophosphatase superfamily. BlastP was conducted to compare the protein sequences of ATPases from Pyrococcus furiosus and the bacteria species which under the clan of N-type ATP pyrophosphatase. The E value obtained from the result are below the E-value that is used as the cut-off-score. The bacteria species are Fusobacteruim nucleatum (1e-22), Caldicellulosiruptor saccharolyticus (1e-13),Campylobacter jejuni (4e-19),Chromobacterium violaceum (1e-21)and Polynucleobacter sp. (1e-24). These E value fall below the lowest E value based on BlastP from Pyrococcus furiosus and Macacca mulatta i.e: 5e-50 and 2e-25 respectively. These bacteria species can not be found in BlastP on the first place because the E value are quite high which indicate the sequence similarity between bacteria species and Pyrococcus furiosus and Macaca mulatta are very low. These suggested that although some bacteria species still have protein sequences which belong to N-type ATP pyrophosphtase superfamily, they are distantly related to both archaea and eukarya. Based on the high protein sequence similarity between Archaea and Eukarya, it can be suggested that Archaea and Eukarya are closely related compared to that for Bacteria.
Based on unrooted tree, the relatedness of the taxa and their relationship can be illustrated but the last common ancestor and how the species evolved cannot be observed. It was found that the taxa are clearly grouped into Domain Archaea and Eukarya. Moreover, in Eukarya, the taxa are nicely clustered according to parasites, vertebrates and invertebrates. Domain Bacteria is missing in this tree suggesting that Bacteria are distantly related to Archaea and Eukarya based on the sequence of ATP Binding Domain 4. Phylogram (figure 3.0) is another way to illustrate the evolution of the taxa where the relationships between the taxa and the time or rate of evolution can be observed.
Bootstrap was conducted to test the reliability of the branching order of the phylogenetic tree. Based on the bootstrap value, we can be confident with the order of the phylogenetic tree thus allowing us to determine where speciation events occur. Bootstrap worked by performing 'pseudoreplicates' of multiple sequence alignments and in this project, 100 replicates was performed. The bootstrap tree can then be generated thus comparing the branching orders and distances of phylogenetic tree. The bootstrap value for each branch are obtained in percentage which indicate the confidence of the branch being correct. If the value of bootstrap is less than 75%, the branching order is not very reliable and meaningless. If the value is between 90% and above, we can be confident that the branching orders are correct. Based on the bootstrap result, it was found that most of the value are lower than 75% suggesting that the branching patterns and distances are not reliable. Therefore, another phylogenetic tree need to be built in order to increase the reliability of the branching pattern and distances of the tree.
Based on unrooted tree, it was found members of Domain Eukarya are nicely cluster at one side without any presence of other species from different domain i.e: members from different domains are well-seperated. Therefore, it can be suggested that the evolutionary model for the PP-loop motif is hold since there is no evidence of the occurrence of lateral gene transfer.
These result support theory for origins of eukaryotes which suggested chimeric features of eukaryote genome (fusion of archaea and bacteria). Study which involved whole-genome-sequence data to test this theory, indicates Eukaryote genome is a chimera of genes most similar to that in Archaea and Bacteria. This study used 'homology-hit' analysis in which the genes from eukaryotes from different classes matching to nearest homology genes in Archaea and Bacteria. It was found that informational genes are closely related to Archaea whereas operational genes are closely related to bacteria (Horiike et al., 2001). Moreover, study conducted by Brown and Doolittle which involved analysing geneolgies from 66 protein-coding genes from members of all three domains of life found that Arginosuccinate synthase from Eukaryotes are closely related to Archaea (analysis on structural similarity found that the conserved region of Arginosuccinate synthase are similar to that of ATP pyrophosphatase)(Katz, 1998). Hence, finding from previous studies support the evolutionary relationship between the three Domain of life based on ATP Binding Domain 4 which suggested that that Archaea and Eukarya are closely related.
Discussion On Structure
ATP binding Domain 4 (IRU8) is a protein with an unknown function. The crystallization of 1RU8 was done from Pyrococcus furiosus that is expressed in Escherichia Coli with resolution of 2.7 Angstroms and an R-value of 0.218. Artificial chemical ligand, TRS(2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL) was used in the crystallization in order to maintain the integrity of the structure when crystallized. There is no actual ligand of this protein based on the the literature paper and from the Protein Data Base (PDB). This is probably because the function of the protein is still unknown. Determination of the ligand can also leads to the possible function of the protein. Hence, deduction of the ligand of this protein can only be done, based on other protein's ligand that has similar function or structure.
From the structure obtained from PyMOL, visualization of the secondary structure provide information on the surface properties covering positively charged regions and negatively charged regions (figure 3.0), its domains (figure 2.0) as well as ligand binding sites and surface clefts (figure 7.0), and also conservation of residues across different species(figure 2.0). It was observed that detailed secondary structure can be determined using PDBsum where conserved residues, protein motif and possible ligand binding site was highlighted (Figure 2.0). Information from the PDB and SCOP has indicated that 1RU8 is from the family of N-type ATP pyrophosphatases and also under the clan of PP-loop which is the strongly conserved motif Ser(12),Gly(13),Gly(14),Lys(15) and Asp(16) at the N terminus (Bork & Koonin 1994). This PP-loop was significant as it is the most conserved and is involved in ligand bindings and thus is likely to give function to our protein. Besides that, from CATH domain database, two main domains were found in 1RU8. The first domain ranges from residue 3-97 while the second domain residues from 98-232. Topology of Domain 1 is known to be of Rossmann A-B-A fold, and the superfamily of PP-loop is presence in this domain. Domain 2 is A-B complex classified in CATH.
Structural alignment via DALI enabled us to obtained proteins that is similar in terms of structure relatedness to 1RU8. Based on the DALI output, 2D13 is rejected since it is a hypothetical protein ph1257 that has no known function to compare to. Thus, Arginosuccinate Synthase (2NZ2) and Queuosine Biosynthesis (3BL5) protein was chosen since it is the most similar based on the z-score (Figure 4.0). Information on 2NZ2 and 3BL5 by InterPro indicated that both protein has conserved motif of PP-loop at the N terminus which is similar to our protein. Hence, emphasizing again the importance of PP-loop to 1RU8's function. These 2 protein were analyzed based on the structural alignment and cleft size and volume. We inferred that the protein that has the most similar features to 1RU8 will probably gives similar function to 1RU8.
Structural alignment of 1RU8 with 2NZ2 and 3BL5 (Figure 5.0 and 6.0) show that 1RU8 and 2NZ2 has the most similar alignment compared to 1RU8 and 3BL5. Since 2NZ2 has a known function which is to catalyzes the citrulline and aspartate into argininosuccinate and pyrophosphate via hydrolysis of ATP, we inferred that the function of our protein may have similar mechanisms to 2NZ2 particularly the hydrolysis of ATP mechanisms. Based on Figure 5.0, we can observed that the ATP is located near the PP-loop highlighted in blue and Citrulline(yellow) is quite far from the loop which is supporting the fact that ATP is hydolysed for citrulline to activate. Hence, similar mechanism may be imply to 1RU8. Besides that, the active site or the binding site structure of both protein is quite similar(based on the structural alignment), and thus we also inferred that the substrate for 1RU8 must then have similar properties and organization of 2nz2 substrate (ATP and Citrulline) in a certain extent. However, the alignment is not convincingly similar due to domain B of 1RU8 (Figure 5.0) that probably is the distinction of differences in function to 2NZ2.
Pockets and cavities in the structure is often associated with binding sites and active sites of proteins. Moreover, it is also believed that there is high possibility that the largest cavity is the active site with some exceptions (Liang et al. 1998). Shape and size parameters of protein pockets and cavities are thus are important for active site analysis. Identification and measurements of surface accessible pockets as well as interior inaccessible cavities of 1RU8, 2NZ2 and 3BL5 were obtained from CASTp (Figure 7.0 to 7.2). It is inferred that the cleft volumes in proteins are related to their molecular interactions and functions (Laskowski et al. 1996). It was observed from the result, cleft of 1RU8 is quite similar in size and volume to 2NZ2 which suggesting that 1RU8 ligand may have similar size to 2NZ2 ligand. Thus, supporting the hypothesis that 1RU8 substrate or ligand may have similar properties to 2NZ2 substrate.
Despite the comparison of the two proteins to 1RU8, further structural comparison is needed in order for us to deduces other possible functions. Besides that determination of actual ligand based on experimental data may also help in finding the function of 1RU8.
Discussion on Function
From our findings, since both argininosuccinate synthetase (AS) and ATP binding domain 4 has striking similarities in terms of highly conserved PP-loop motif and cleft volume, it is possible to infer the mechanism of our protein based on the well-known AS reaction mechanism.
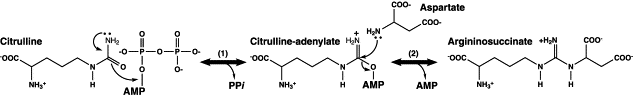
From the figure 8.0,
Step 1. Argininosuccinate synthetase releases inorganic pyrophosphate after the formation of activate citrulline-adenylate.
Step 2. Aspartate (the lone pair of N from amino group) undergoes nucleophilic attack on the carbonyl group on the activated citrulline-adenylate, hence forming argininosuccinate together with the release of AMP.
Therefore,
(1) Structure modelling with argininosuccinate synthetase (AS).
Our protein should be also involved in catalysing a substrate adenylation (means forming a phosphodiester bond between an amino acid and the phosphate group of AMP (adenosine monophosphate nucleotide) or simply the process of forming an adenylate, the salt or an ester of AMP, so as to activate a carbonyl or carboxyl group. (Lemke & Howell 2001) This activation is to facilitate the subsequent attack of a nitrogen-containing nucleophile from the substrate.
The structure of AS is used for the modelling ATP binding into our structure. The beta and gamma phosphate groups of ATP are oriented by characteristic residues of the PP-loop, and then forming a salt linkage between the g-phosphate and the N atom of lysine. (Lemke & Howell 2001)
In the AS model in E. coli., R168 (see figure 8.1) has been proposed for pyrophosphate binding. The carbonyl oxygen of the second residue preceding the PP motif in AS (Ala-16), falls within the hydrogen bonding distance of the O2’ hydroxyl oxygen of the ribose. (Lemke & Howell 2001). Therefore, the alanine residue closely after the PP-motif in our protein, Ala(20) should be involved in stabilising ribose by forming H-bond with its O2’ hydroxyl oxygen. This may explain why relatively majority of aliphatic residues are conserved in the biggest gap in our cleft analysis (see figure 4.2 under function analysis) and relatively highly conserved detected by PDBsum.
It was proposed that the amide oxygen of a glutamine (Q46) should be involved in interacting with the N6 nitrogen of adenine. Relatively highly conserved Q104 in our structure (detected by PDBsum) might also be involved in similar interaction.
The most intriguing part is in the highly conserved PP-loop motif ([A/S]-[F/Y]-S-G-G-[L/V]-D-T-[S/T]) contains two absolutely conserved glycine residues, Gly(13) and Gly(14). In AS model, the two conserved glycine residues showed significantly different conformations in its uncomplexed and complexed structures, which are suspected to play a role in binding and release of pyrophosphate (PPi). (Lemke & Howell 2001) This agrees with glycine, lacking side chains, can provide a high degree of flexibility. Therefore, Gly(13)Gly(14) in ATP binding domain 4 should provide a large anion hole required for pyrophosphate binding. Lemke and Howell (2001) proposed that other N-type ATP pyrophosphatases models, if having glycine residues replaced, the steric hindrance created will crash with the bridge oxygen of the bound pyrophosphate.
The final residue of the PP motif in our structure, the hydroxyl group of Ser(17) should be involved in forming hydrogen bonds with the highly conserved residues, Gly(14) and Arg(60), meaning Ser(17) residue is important both for pyrophosphate binding and structural support by interacting with a helices, therefore H1 and H2 can be linked together. (see Figure 2.0 showing H1 and H2 in PDBsum)
(2) Protein conformational change existed in the catalytic cycle.
The relative positions of all 3 substrates in AS suggest a strong requirement for a conformational change during catalysis. Our analysis of AS with substrates interactions indicates that PP-loop closely binds with ATP and is distantly related to citrulline (the substrate). This implies that as for our protein, only PP-loop is directly interacting with ATP, and its substrate may interact with ATP in the opposite end.
(3) Only domain A was involved in ATP-substrate binding; domain B suggested functions otherwise.
Domain B in our protein is poorly aligned with AS but closely resembled only in PP-loop-containing domain A region, which has been previously confirmed in our PyMOL alignment. It suggested that domain B of ATP binding domain 4 may be involved in substrates recognition which explains the subtle differences between our protein with AS. This hypothesis was based on our findings that the starting residues of domain B formed a hairpin-like structure that pointed to the core of ATP binding domain 4 that might suggest an intermolecular cooperation.
Future research should be focused on developing the crystallography of ATP binding domain 4 binding with its substrates. Attempts of building a cocrystallized structure with ATP and enzyme is often hard. Taking Lemke and Howell (2001)’s study as an example, cocrystallizing ATP with AS from E. coli. is detrimental which results into a poorly diffracting, easily cracked and dissolving crystals. Therefore, our study focuses only on inferring structure based on other N-type ATP pyrophosphatases. However, building a crystallised model will definitely provide us with insight to the enzymatic mechanism.
More structural and functional deduction experiments should be done based on site-directed mutagenesis on PP-loop and potential substrate-interacting residues in the groove identified in our structural analysis. More studies should be done based on elucidating domain B of ATP binding domain 4 which may be important for substrate selectivity.
Further research can be done on comparing the ATP binding domain 4 between human and Pyrococcus furiosus and look for any mutations of the structurally and catalytically important residues suggested in this study are associated with any ATP-related genetic diseases.
Abstract|
Introductions|
Methods|
Structural Analysis|
Functional Analysis|
Evolutionary Analysis|
Discussions|
Conclusion |
References