C1orf41 Results: Difference between revisions
No edit summary |
No edit summary |
||
| (30 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
'''Structure''' | '''Structure''' | ||
PDB ID: 1TVG | |||
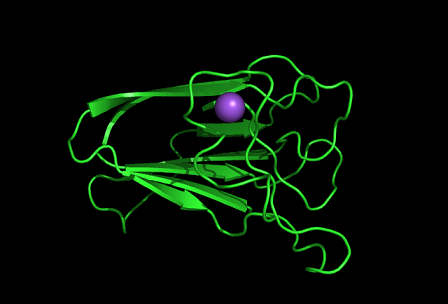
Information from the PDB stated that x-ray diffraction was ueed to solve the structure of this protein with an R value of 0.215 and at 1.6Å. Two ligands were present in the crystal structure, a calcium (II) ion and a samarium (III) ion. This protein has 153 residues and its secondary structure consists of two helices and eight beta strands. | |||
Scop result | Scop result | ||
| Line 11: | Line 10: | ||
[[Image:PDB sum 2.PNG | [[Image:PDB sum 2.PNG|centre|framed|'''Figure 1:''' Representation of c1orf41 secondary structure as presented in PDBsum.]] | ||
Figure 1: | |||
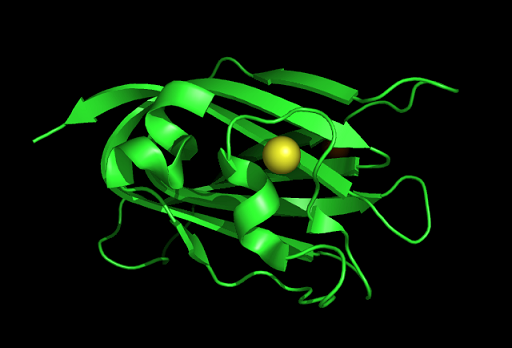
Tertiary structure | Tertiary structure | ||
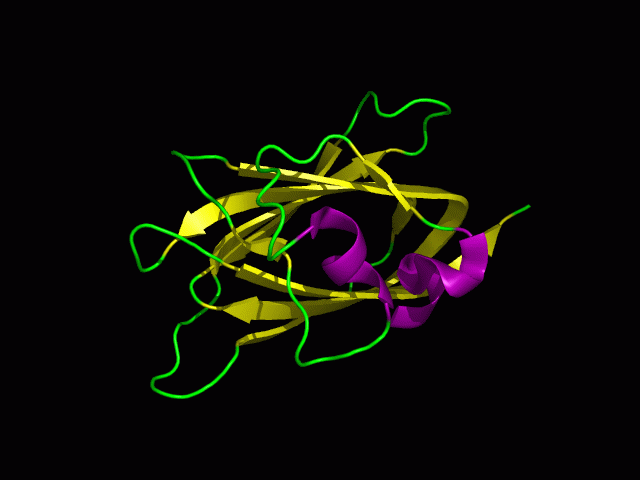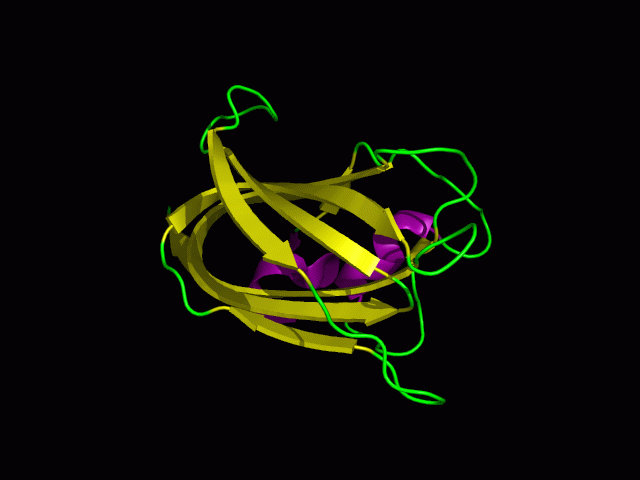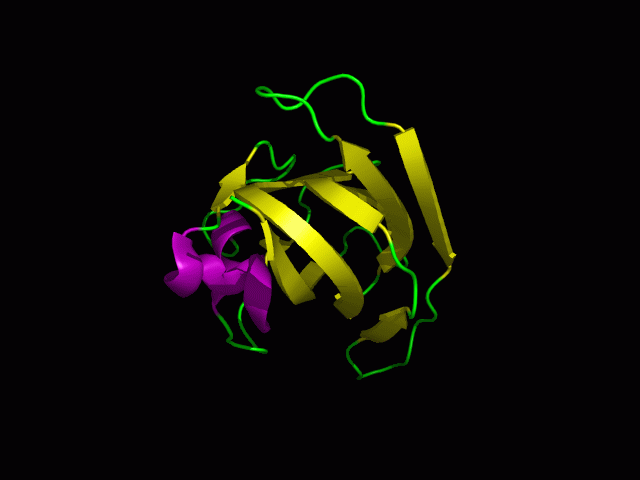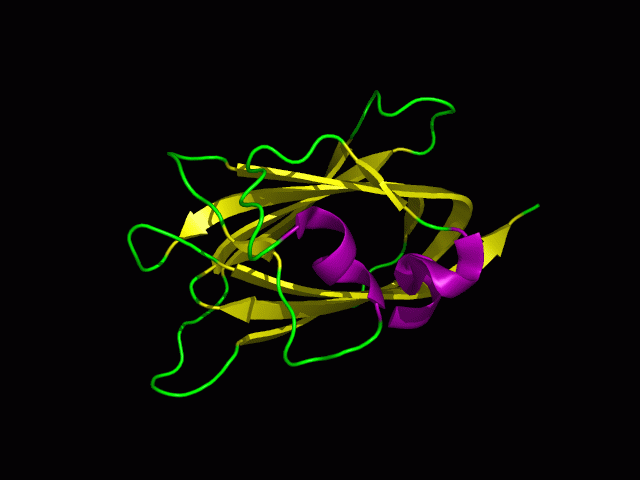
[[Image:1tvg ani.gif]] | [[Image:1tvg ani.gif|centre|framed|'''Figure 2:''' This protein is monomeric, consisting of a single domain. There are 8 beta strands (yellow) and 2 alpha helices (purple). The secondary structures are folded into jelly roll barrel where four pairs of anti-parallel beta strands are organised to form a barrel-like structure.]] | ||
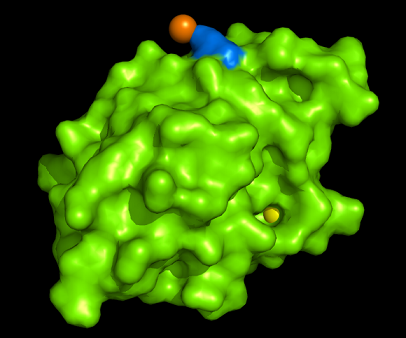
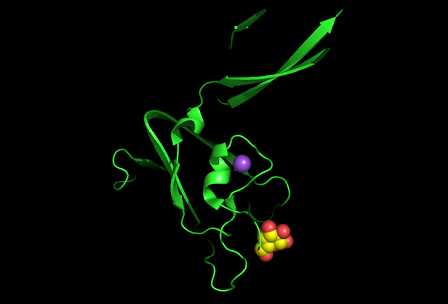
[[Image:1tvg surface.png|centre|framed|'''Figure 3:''' Surface representation of c1orf41. The Ca (II) ion is located within a pocket of the protein. Sm (III) interacts with Asp92 on the protein but its presence was probably due to the method used in solving the phase problem during structure solution using x-ray crystallogrophy.]] | |||
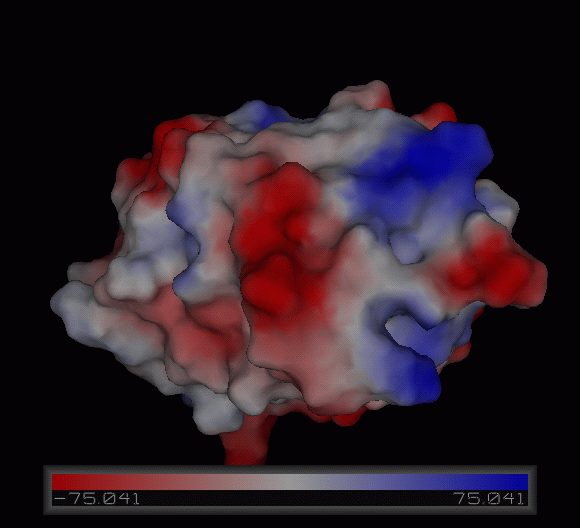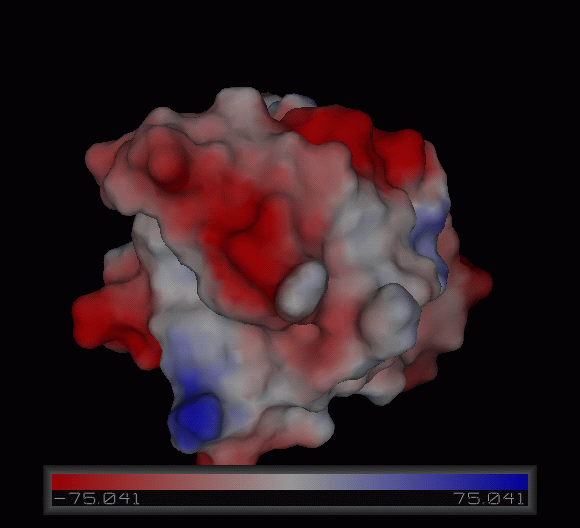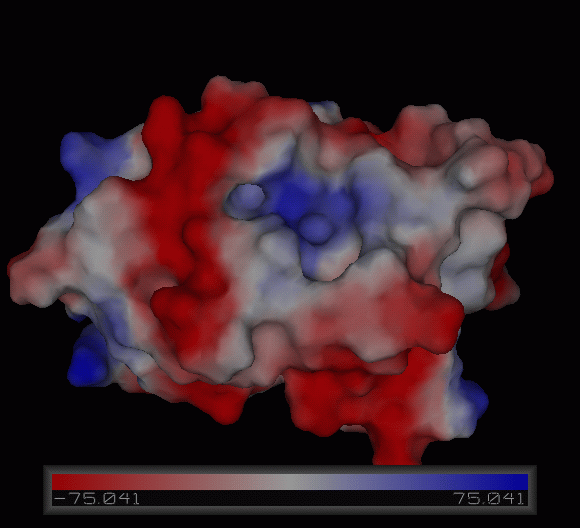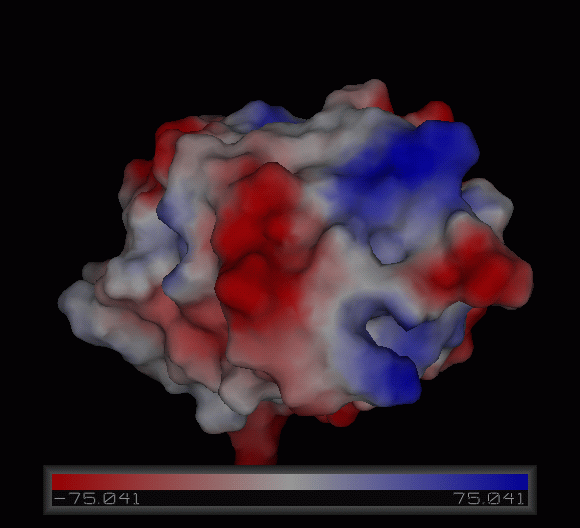
Figure | [[Image:Charge surface ani.gif|centre|framed|'''Figure 4:''' Electrostatic charge distribution on the surface of c1orf41.]] | ||
---- | ---- | ||
''' | '''Structural analysis''' | ||
'''1)Structural similarities''' | |||
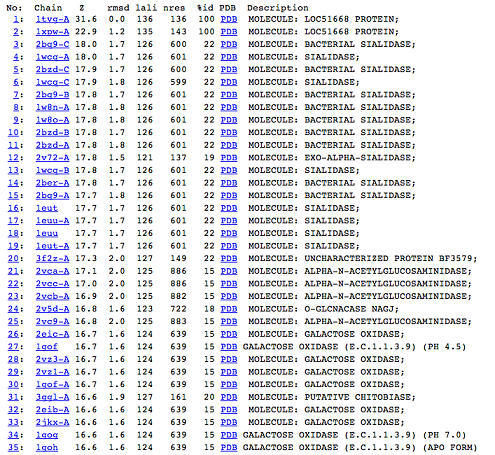
DALI was used to find proteins that are similar in structure to c1orf41. The results showed that sialidases, alpha-N-acetylglucosaminidases and galactose oxidases have similar structures to this protein. | |||
DALI | [[Image:Picture 1.png|centre|framed|'''Figure 5:''' DALI result showing the first 35 proteins that have similar structures to c1orf41. Even though the proteins are similar in structure they have very little sequence identity to c1orf41. 1xpw is c1orf41 but the structure was solved by NMR.]] | ||
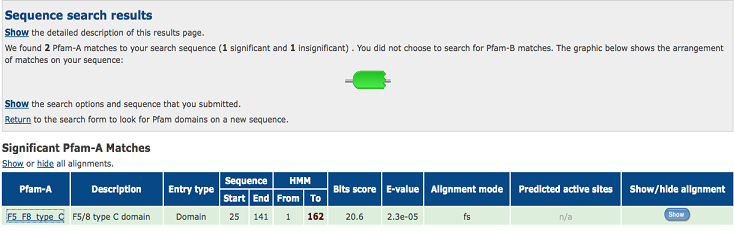
'''2)Domain Classification''' | |||
[[Image:1TVG pfam.png|centre|framed|'''Figure 6:''' Pfam result indicated that our protein has a F5/8 type C domain, also known as the discoidin domain which is apart of galactose binding domain super family.]] | |||
'''3)Possible ligand binding sites''' | |||
Observations and comparisons of several proteins from the DALI result with our protein showed that there is a similar position on each proteins where a metal ion is located. | |||
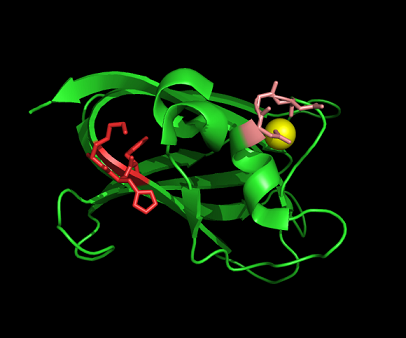
[[Image:1tvg ligand1.png|centre|framed|'''Figure 7:''' This figure shows the position where calcium (II) ion (yellow) is located in within a loop of c1orf41 structure. This loop where metal ion is coordinated to was also observed in several proteins from the DALI result.]] | |||
[[Image:1gog Na.png|centre|framed|]] | |||
[[Image:2bzd discoidin2.png|centre|framed|'''Figure 8:'''Shown at the top is the discoidin domain of galactose oxidase with a sodium ion (purple) located within a loop. The bottom figure is bacterial sialidase discoidin domain with a sodium ion (purple) and a beta-D-galactose molecule. The sodium ion is also located i within a loop.]] | |||
Figure | [[Image:1tvg castp3.PNG|centre|framed|'''Figure 9:'''CastP identified a surface cleft that is located at the loop position.]] | ||
[[Image: | [[Image:1tvg ca res2.PNG|centre|framed|'''Figure 10:'''Pymol figure displaying possible metal binding position on c1orf41. Pink are the residues that may be involved in coordinating the metal and yellow is the calcium ion. The residues are based on CastP result. The aspartate and glutamate residues are likely to be important in binding positively charged metal ion]] | ||
Nest analysis by Profunc suggested two other ligand binding sites. Nests are structural motifs that are commonly found in functionally important regions of protein structures. | |||
[[Image:Profunc nest.PNG|centre|framed|'''Figure 11:'''Nest 1 and 2 had scores higher than 2. So, they are more likely to be functional. His54 and Lys55 NH atoms are accessible from a large surface cleft of the protein. The cleft is also deep indicating that the nest is functionally important. The residues of Nest 2 are part of the loop that coordinates the Ca (II)ion.]] | |||
[[Image:1tvg | [[Image:1tvg nests2.png|centre|framed|'''Figure 12:'''Pymol representation of Nest 1 and 2.]] | ||
[[Image:Surface nest3.PNG|centre|framed|'''Figure 13:'''Arrows indicating the pockets of the nests that may be accessible to ligands.]] | |||
---- | |||
'''Evolution''' | |||
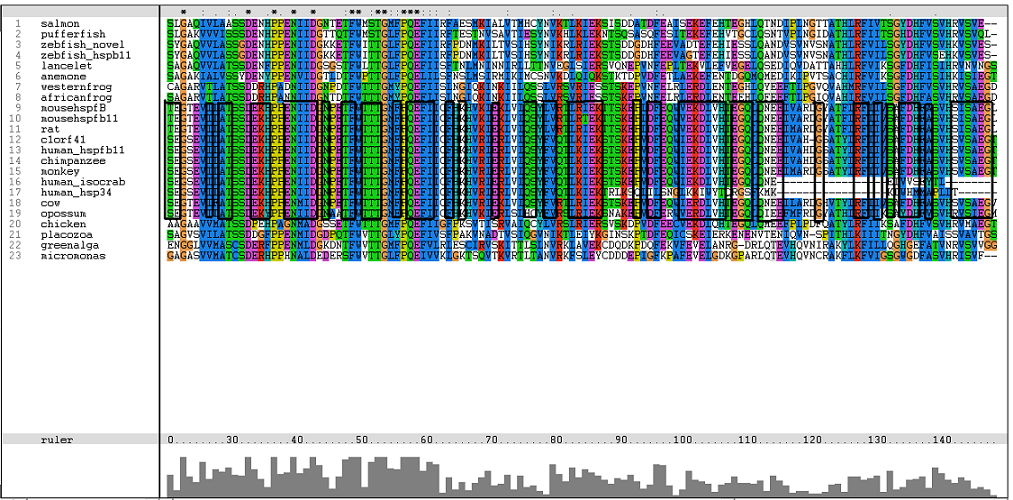
Results from Blast showed that c1orf41 have high homology to small heat shock proteins which gave us a hint that the protein may be a heat shock protein (Figure 14). However, initial multiple sequence alignment showed many gaps and few conserved regions (Figure 15). | |||
[[Image:BLAST.png|centre|framed|'''Figure 14:''' Homologues of c1orf41 from BLAST.]] | |||
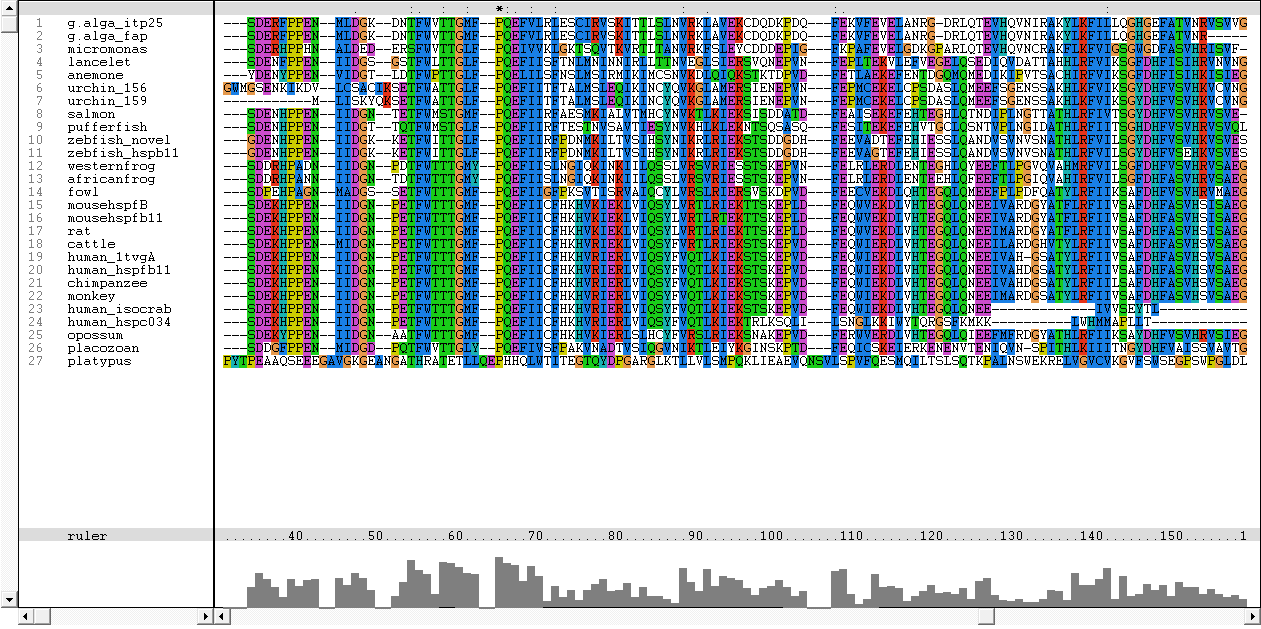
Figure | [[Image:Platypus.png|centre|framed|'''Figure 15:''' Initial multiple sequence alignment of the 27 proteins obtained from Blast]] | ||
The sequences from platypus, urchin_156, urchin_159 and g.alga_fap were excluded from further investigation. This was due to poor alignment of platypus, urchin_156 and urchin_159 with the other organisms. In addition, the sequence for g.alga_fap was removed as it was a truncated sequence of g.alga_itp25. | |||
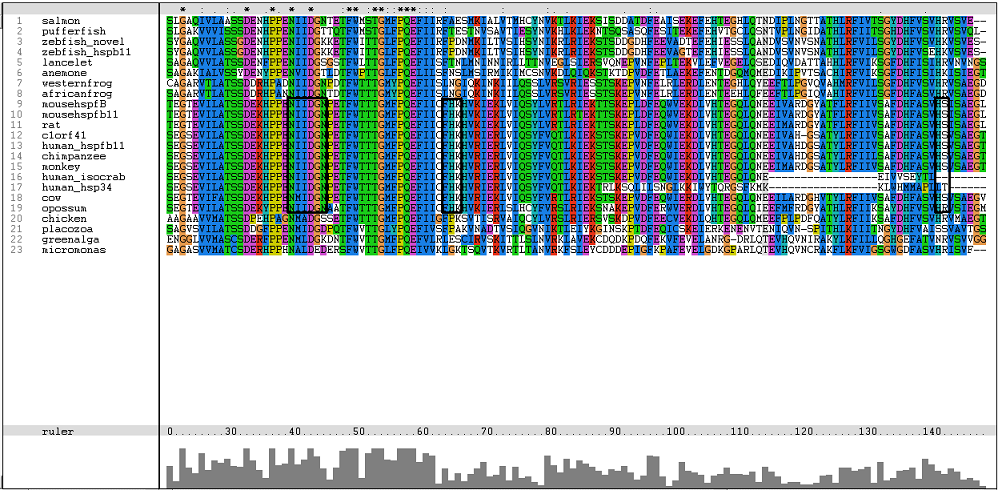
One of the characteristic of a small heat shock protein is that they have a conserved alpha B crystallin domain which can also be observed in c1orf41 as shown in figure 16 below: | |||
Figure | [[Image:Msa16.png|centre|framed|'''Figure 16:''' Conserved alpha B crystallin domain in c1orf41]] | ||
The conserved residues found in c1orf41 and its homologues fall in the alpha B crystallin domain. But more conserved regions are observed in higher terrestrial eukaryotes. With this, we are more confident that c1orf41 is a heat shock protein. | |||
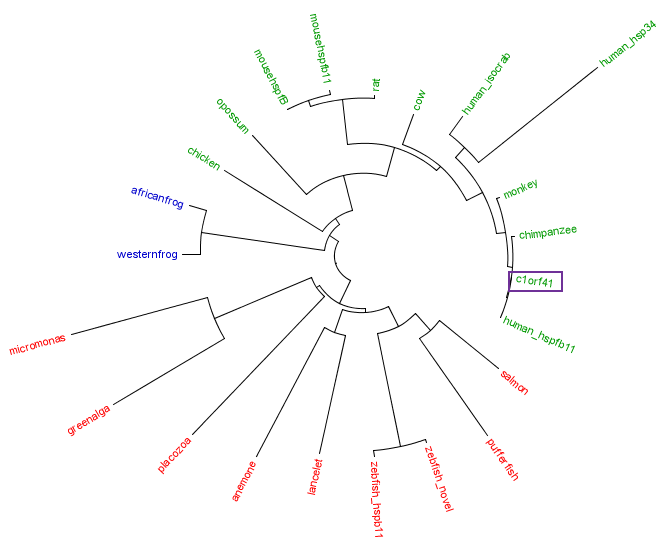
The residues of the possible binding site were highlighted in black in figure 17 below. However, these residues were only conserved in terrestrial eukaryotes. During evolution, the protein has evolve and adopt different function in marine eukaryotes, amphibian and terrestrial eukaryotes. The evolutionary relationship of this protein is shown in figure 18. | |||
[[Image:Msa17.png|centre|framed|'''Figure 17:''' Residues involved in the possible binding site of c1orf41]] | |||
[[Image: | [[Image:Treecolour.png|centre|framed|'''Figure 18:''' Unrooted phylogenetic tree of c1orf41 and its homologues. The tree showed that the protein evolved from marine eukaryotes (red) to amphibians (blue) and to terrestrial eukaryotes (green)]] | ||
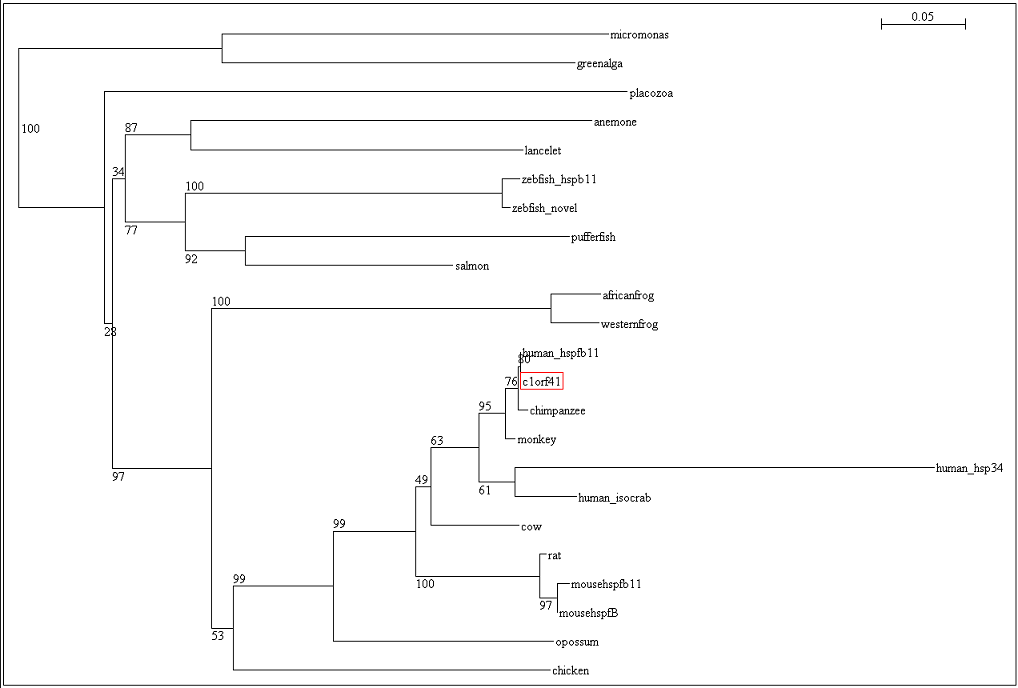
To ensure that the reliability of the evolutionary relationship of c1orf41 and its homologues, the tree was bootstrap with 100 replicates (Figure 19). Bootstrap value of more than 75 indicated that we are confident in the branch whereas bootstrap value of less than 50 indicated no confidence in the branch over an alternative | |||
[[Image:Tree.png|centre|framed|'''Figure 19:''' Phylogenetic tree with its respective bootstrap value]] | |||
---- | |||
'''Function''' | |||
Information from sequence analysis (i.e. Blast, structure analysis (i.e.domain classification (Pfam) and possible ligand binding site) was used to infer the function of c1orf41. Profunc server was also run to get a summary of sequence and structural analysis. Relevant information was used to analyze the function. | |||
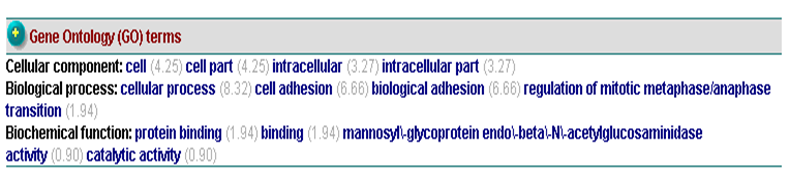
[[Image: | Gene ontology terms suggested that c1orf41 involves in cellular process or cell adhesion (Figure 20). | ||
[[Image:GO.png|centre|framed|'''Figure 20:''' Gene ontology terms of c1orf41.]] | |||
Based on Blast result, our protein has high similarity with sHsp. Thus, they may have similar function. sHsp functions as molecular chaperone and cytotprotective (antiapoptotic). The C1orf41 has a loop binds to Ca2+ which presumably related to antiapoptotic function of sHsp. | |||
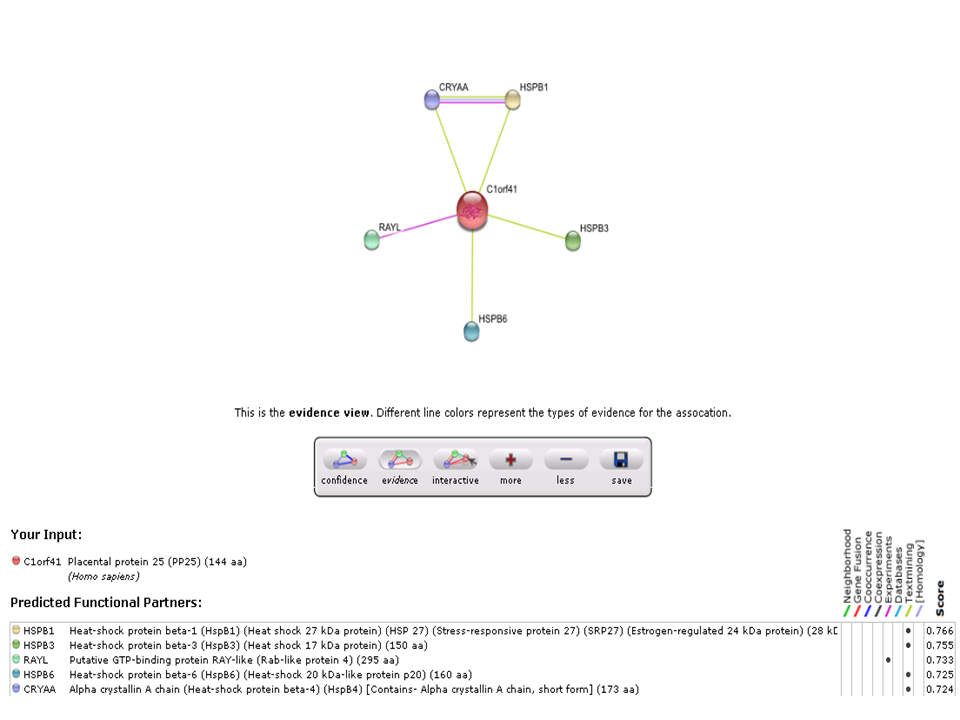
The String analysis showed protein-protein interactions of c1orf41. Based on experimental evidence, there was an interaction between c1orf41 and RAYL (Ras-like-GTPase superfamily member). Overexpression of Ras GTPase activating protein will enhance the phosphorylation & activity of Akt which leads to antiapoptotic activity. | |||
[[Image:string-evidence view.png|centre|framed|'''Figure 21:''' Protein interaction of c1orf41.]] | |||
'''Discoidin''' | |||
Based on Pfam result (Fig.6), our protein target is a member of F5/8 type C (discoidin)domain. It is reported that discodin is involved in cell-cell interaction & recognition and also highly express in breast carcinoma cell lines. However, discoidin is extracellular protein, while c1orf41 is intracellular. Moreover, based on sequence and structural analyses, our protein is more likely a sHsp. | |||
Latest revision as of 01:59, 16 June 2009
Structure
PDB ID: 1TVG Information from the PDB stated that x-ray diffraction was ueed to solve the structure of this protein with an R value of 0.215 and at 1.6Å. Two ligands were present in the crystal structure, a calcium (II) ion and a samarium (III) ion. This protein has 153 residues and its secondary structure consists of two helices and eight beta strands.
Scop result
Tertiary structure
Structural analysis
1)Structural similarities
DALI was used to find proteins that are similar in structure to c1orf41. The results showed that sialidases, alpha-N-acetylglucosaminidases and galactose oxidases have similar structures to this protein.
2)Domain Classification
3)Possible ligand binding sites
Observations and comparisons of several proteins from the DALI result with our protein showed that there is a similar position on each proteins where a metal ion is located.
Nest analysis by Profunc suggested two other ligand binding sites. Nests are structural motifs that are commonly found in functionally important regions of protein structures.
Evolution
Results from Blast showed that c1orf41 have high homology to small heat shock proteins which gave us a hint that the protein may be a heat shock protein (Figure 14). However, initial multiple sequence alignment showed many gaps and few conserved regions (Figure 15).
The sequences from platypus, urchin_156, urchin_159 and g.alga_fap were excluded from further investigation. This was due to poor alignment of platypus, urchin_156 and urchin_159 with the other organisms. In addition, the sequence for g.alga_fap was removed as it was a truncated sequence of g.alga_itp25.
One of the characteristic of a small heat shock protein is that they have a conserved alpha B crystallin domain which can also be observed in c1orf41 as shown in figure 16 below:
The conserved residues found in c1orf41 and its homologues fall in the alpha B crystallin domain. But more conserved regions are observed in higher terrestrial eukaryotes. With this, we are more confident that c1orf41 is a heat shock protein.
The residues of the possible binding site were highlighted in black in figure 17 below. However, these residues were only conserved in terrestrial eukaryotes. During evolution, the protein has evolve and adopt different function in marine eukaryotes, amphibian and terrestrial eukaryotes. The evolutionary relationship of this protein is shown in figure 18.
To ensure that the reliability of the evolutionary relationship of c1orf41 and its homologues, the tree was bootstrap with 100 replicates (Figure 19). Bootstrap value of more than 75 indicated that we are confident in the branch whereas bootstrap value of less than 50 indicated no confidence in the branch over an alternative
Function
Information from sequence analysis (i.e. Blast, structure analysis (i.e.domain classification (Pfam) and possible ligand binding site) was used to infer the function of c1orf41. Profunc server was also run to get a summary of sequence and structural analysis. Relevant information was used to analyze the function.
Gene ontology terms suggested that c1orf41 involves in cellular process or cell adhesion (Figure 20).
Based on Blast result, our protein has high similarity with sHsp. Thus, they may have similar function. sHsp functions as molecular chaperone and cytotprotective (antiapoptotic). The C1orf41 has a loop binds to Ca2+ which presumably related to antiapoptotic function of sHsp.
The String analysis showed protein-protein interactions of c1orf41. Based on experimental evidence, there was an interaction between c1orf41 and RAYL (Ras-like-GTPase superfamily member). Overexpression of Ras GTPase activating protein will enhance the phosphorylation & activity of Akt which leads to antiapoptotic activity.
Discoidin
Based on Pfam result (Fig.6), our protein target is a member of F5/8 type C (discoidin)domain. It is reported that discodin is involved in cell-cell interaction & recognition and also highly express in breast carcinoma cell lines. However, discoidin is extracellular protein, while c1orf41 is intracellular. Moreover, based on sequence and structural analyses, our protein is more likely a sHsp.