Chromosome 1 open reading frame 41 Structure: Difference between revisions
Hana Hamzah (talk | contribs) No edit summary |
Hana Hamzah (talk | contribs) No edit summary |
||
| Line 61: | Line 61: | ||
Figure 7: This figure shows the position where calcium (II) ion (yellow) is located in within a loop of c1orf41 structure. This loop where metal ion is coordinated to was also observed in several proteins from the DALI result. | Figure 7: This figure shows the position where calcium (II) ion (yellow) is located in within a loop of c1orf41 structure. This loop where metal ion is coordinated to was also observed in several proteins from the DALI result. | ||
[[Image:1gog Na.png]] | [[Image:1gog Na.png]] | ||
| Line 73: | Line 74: | ||
Figure 9: CastP also identified a surface cleft that is located at the loop position. | Figure 9: CastP also identified a surface cleft that is located at the loop position. | ||
[[Image:1tvg | [[Image:1tvg ca res2.PNG]] | ||
Figure 10: Pymol figure displaying possible metal binding position on c1orf41. Orange is the residues that may be involved in coordinating the metal and yellow is the calcium ion. The residues are based on CastP result. | |||
Nest analysis by Profunc suggested two other ligand binding sites. | Nest analysis by Profunc suggested two other ligand binding sites. Nests are structural motifs that commonly found in functionally important regions of protein structures. | ||
[[Image:Nest analysis result.PNG]] | [[Image:Nest analysis result.PNG]] | ||
Figure 11: Nest 1 and has scores higher than 2. So, they are more likely to be functional. His54 and Lys55 NH atoms are accessible from a large surface cleft of the protein. The cleft is also deep indicating that the nest is functionally important. The residues of Nest 2 are part of the loop that coordinates the Ca ion. | |||
[[Image:1tvg | [[Image:1tvg nests2.png]] | ||
Revision as of 23:42, 9 June 2009
Protein Structure
PDB ID: 1TVG Information from the PDB stated that x-ray diffraction was sued to solve the structure of this protein with an R value of 0.215 and at 1.6Å. Two ligands were present in the crystal structure, a calcium (II) ion and a samarium (III) ion. This protein has 153 residues and its secondary structure consists of two helices and nine beta sheet strands.
Scop result Root: scop Class: All beta proteins [48724] Fold: Galactose-binding domain-like [49784] sandwich; 9 strands in 2 sheets; jelly-roll Superfamily: Galactose-binding domain-like [49785] uperfamily Family: APC10-like [69210] Pfam 03256 Protein: Placental protein 25, pp25 [117104] Species: Human (Homo sapiens) [TaxId: 9606] [117105] SQ Q9Y547
Figure 1: Above is the representation of c1orf41 secondary structure as presented in PDBsum.
Tertiary structure
Figure 2: This cartoon representation of c1orf41 shows that this protein is monomeric, consisting of a single domain. the fold of the protein is a jelly roll barrel where four pairs of anti-parallel beta strands are organised to form a barrel-like structure.
Structure analysis
1)Structure similarities
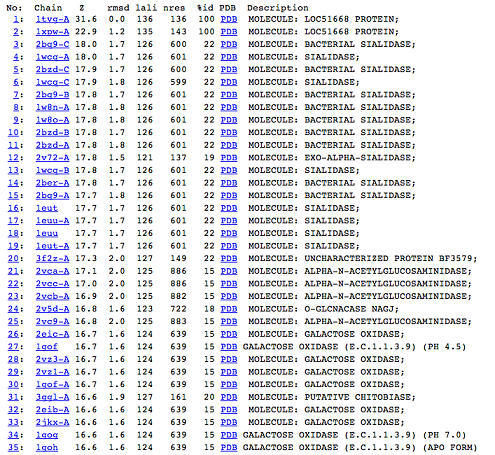
DALI was used to find proteins that are similar in structure to c1orf41. The results showed that sialidases, alpha-N-acetylglucosaminidases and galactose oxidases have similar structure to this protein.
Figure 5: DALI result showing the first 35 proteins that have similar structures to c1orf41. Even though the proteins are similar in structure they have very little sequence identity to c1orf41. 1xpw is c1orf41 but the structure was solved by NMR.
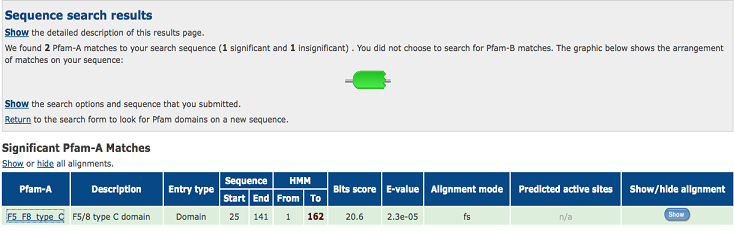
2)Domain Classification
Figure 6:Pfam result indicated that our protein has a F5/8 type C domain, also known as the discoidin domain which is apart of galactose binding domain super family.
Closer inspection of several of the other proteins from the DALI result showed that they also have discoidin domain in their structures. It was probably based on this domain that the DALI result was obtained.
3)Possible ligand binding sites
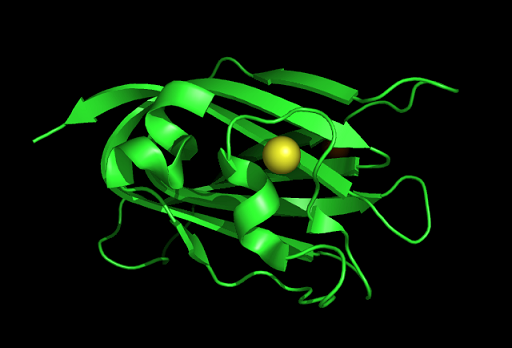
Observations and comparisons of several proteins from the DALI result with our protein showed that there is a similar position on each proteins where a metal ion is located.
Figure 7: This figure shows the position where calcium (II) ion (yellow) is located in within a loop of c1orf41 structure. This loop where metal ion is coordinated to was also observed in several proteins from the DALI result.
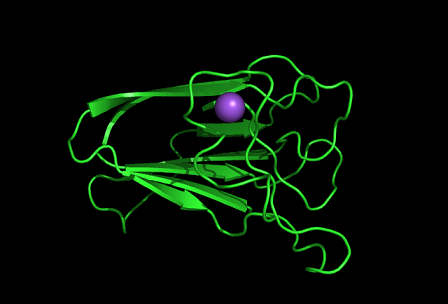
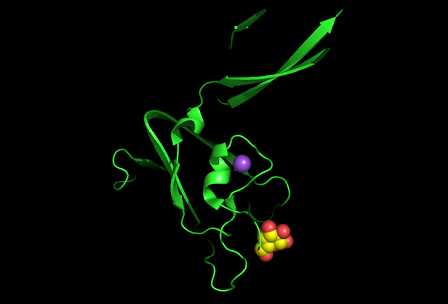
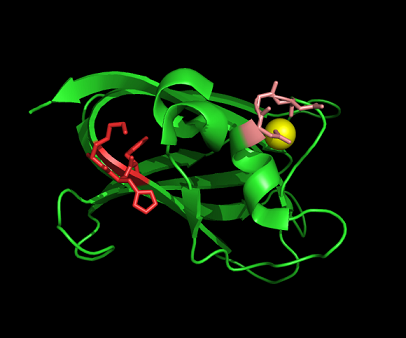
Figure 8: Shown at the top left is the discoidin domain of galactose oxidase with a sodium ion (purple) located within a loop. The bottom figure is bacterial sialidase discoidin domain with a sodium ion (purple) and a beta-D-galactose molecule. The sodium ion is also located i within a loop.
Figure 9: CastP also identified a surface cleft that is located at the loop position.
Figure 10: Pymol figure displaying possible metal binding position on c1orf41. Orange is the residues that may be involved in coordinating the metal and yellow is the calcium ion. The residues are based on CastP result.
Nest analysis by Profunc suggested two other ligand binding sites. Nests are structural motifs that commonly found in functionally important regions of protein structures.
Figure 11: Nest 1 and has scores higher than 2. So, they are more likely to be functional. His54 and Lys55 NH atoms are accessible from a large surface cleft of the protein. The cleft is also deep indicating that the nest is functionally important. The residues of Nest 2 are part of the loop that coordinates the Ca ion.