Chromosome 1 open reading frame 41 Structure
Protein Structure
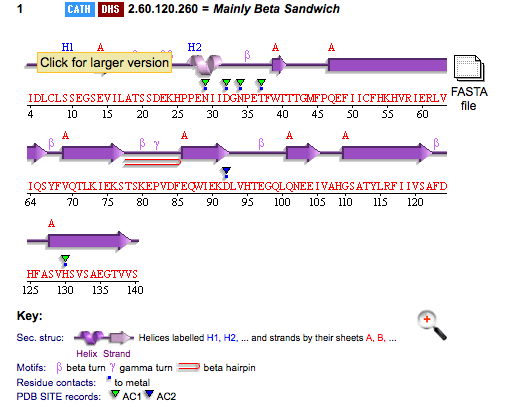
PDB ID: 1TVG Information from the PDB stated that x-ray diffraction was sued to solve the structure of this protein with an R value of 0.215 and at 1.6Å. Two ligands were present in the crystal structure, a calcium (II) ion and a samarium (III) ion. This protein has 153 residues and its secondary structure consists of two helices and nine beta sheet strands.
Above is the representation of 1TVG secondary structure using PDBsum.
This protein is monomeric and the connectivities between the secondary structures is shown below.
Structure analysis
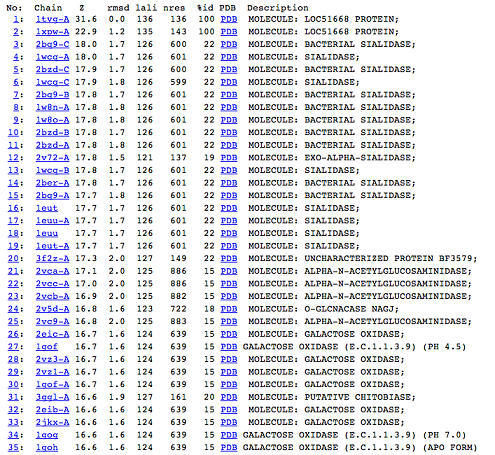
DALI was used to find protein that are similar in structure to 1TVG. The results showed that sialidases, alpha-N-acetylglucosaminidases and galactose oxidases have similar structure to our protein as presented below.
Pymol structure displaying the folding of the secondary structure. Red is alpha helices, yellow is beta sheets and green is random coils connecting the the sheets and helices.