DAP Functional Analysis

The function of DAP was found by researching the few scientific journals on protein as well as analysing the crystal structure in PyMol. The predicted residues were found by matching residues derived experimentally in the human homologue of the protein to P. aeruginosa DAP. The eight Histidine residues 82, 156, 401, 313, 21, 310, 320, & 324 had been given predicted function, however the original authors could not officially assign these roles till the crystal structure had been resolved. Now having the crystal structure of Pseudomona aeruginosa some of these predictions can be validated based on the location of the residues within the protein. The residues are assumed to have a similar function due to the extremely high conservation between species as demonstrated in figure 4.7.
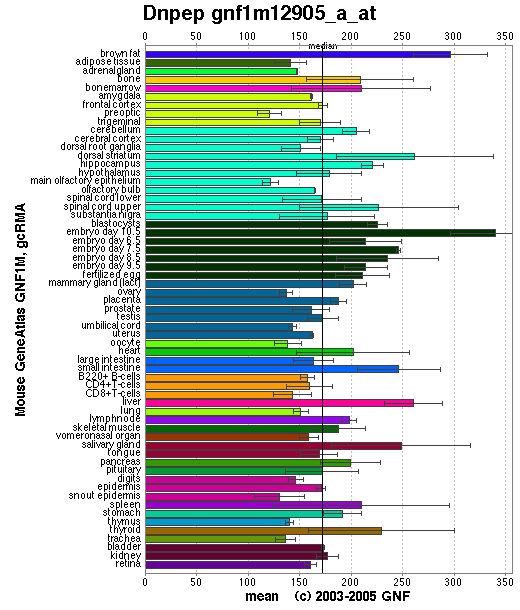
DAP is expressed in teh cytosol of the cell at >0.1% of soluble protein, Wilk et al. (1998). As can be seen in figure 4.2 aspartyl amino peptidase is expressed fairly evenly in mouse tissue. DAP is considered to be a basic metabolic protein degradation enzyme in the cell, and this uniform expression complies with that assesment.
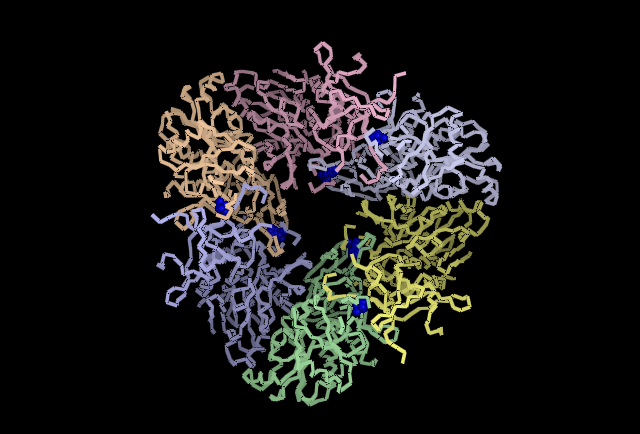
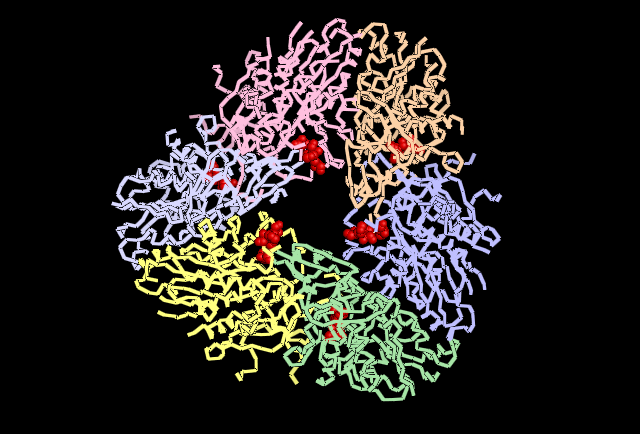
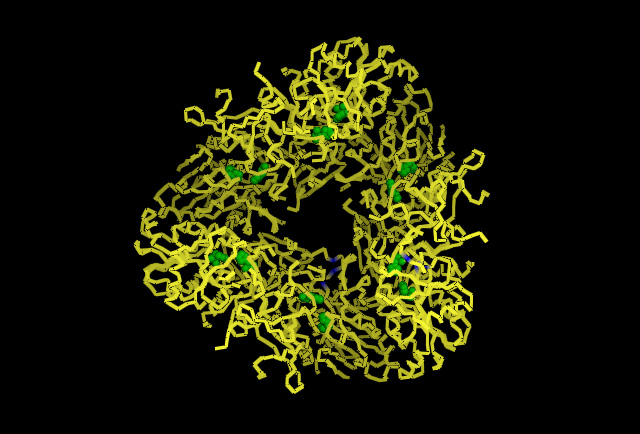
Analysing the first subunit Chain A in PyMol, figure 4.3, it can be seen that the two predicted catalytic His residues 82 and 401 are localised at the centre of the subunit whereas the other predicted residues such as those that reduce kcat (310, 320, &324)and one that is a predicted subunit linking molecule, 313, are located in distincly different areas. This gives the impression they perform different functions. Also highly conserved Asp and Glu from Wilk et al. (1998) was used, and it can be seen in the figure in red that these residues are localised around the predicted catalytic His residues indicating some may have direct involvement in catalytic activity. When the protein is analysed as a full structure the predicted roles of the His residues can be verified.The overall structure is composed of 12 separate subunits and they arrange into a tetrahedral shape, so the protein can be considered to be dodecameric tetrahedral in shape. Also the protein features a channel in the centre of each triangular face. As seen in figure 4.4 His 313 which is believe to be involved in subunit subunit interactions is located directly between adjacent subunits, putting it in a prime position to fulfil that role. In figure 4.5 histidine residues shown to reduce kcat (catalytic constant) are located around the central channel on each triangular surface of the structure, they were previously predicted to have an important structural role in maintiang the shape and integrity of the protein, however based on their location and their function in reducing kcat a role in guiding the substrate to the catalytic centre of the protein is now being proposed. Figure 4.6 demonstrates taht the residues 82 and 401 predicted to be involved in binding zinc to the catalytic centre are located within the tetrahedral structure giving it a catalytic centre, indicating the protein functions similarly to the barel shape observed in other proteins where the substrate specificity is determined by its ability to enter the channels and interact with the active site within.
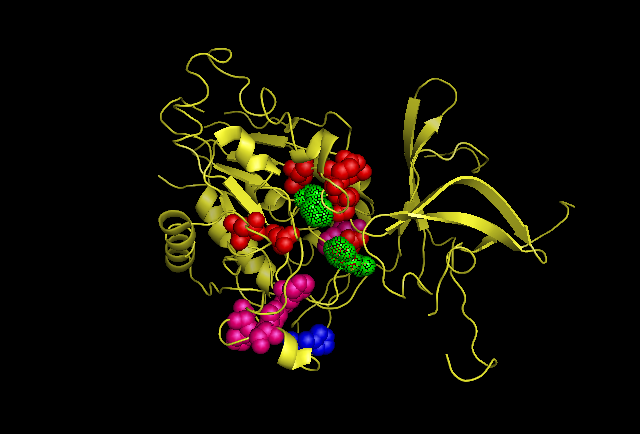
Fig 4.3. This image is chain A. Green shows the predicted active site residues His82 and His401. Pink shows Histidine residues that are also necessary for optimal enzymatic function, and experimentally mutations greatly reduce kcat. Blue is His313 predicted to be involved in subunit-subunit interactions. Red are Aspartyl and Glutamyl residues which are highly conserved across species and thought to participate in catalytic activity.
[2]Return to aspartyl aminopeptidase