DAP Functional Analysis: Difference between revisions
From MDWiki
Jump to navigationJump to search
No edit summary |
No edit summary |
||
| Line 7: | Line 7: | ||
[[Image:possiblecatalyticresidues.png]] | [[Image:possiblecatalyticresidues.png]] | ||
The images below come from UniProt which correctly identifies the protein according to its clan, family and type. As well as predicting very accurately the active site residues corresponding to those found in [Wilk et al. (2002)] | |||
[[Image:uniprotname.jpg]] | [[Image:uniprotname.jpg]] | ||
[[Image:uniprotsequence.jpg]] | [[Image:uniprotsequence.jpg]] | ||
[[Image:uniprotontology.jpg]] | |||
[[Image:prositescan.png]] | [[Image:prositescan.png]] | ||
Fig X. This ProSite scan shows a list of possible active sites with a high probablility of occurence. However they do not seem relevant as they do not take into account the nature of the protein as a metallopeptidase and very few of the predicted catalytic residues feature in this output. | |||
Revision as of 10:03, 7 June 2008
The images below come from UniProt which correctly identifies the protein according to its clan, family and type. As well as predicting very accurately the active site residues corresponding to those found in [Wilk et al. (2002)]
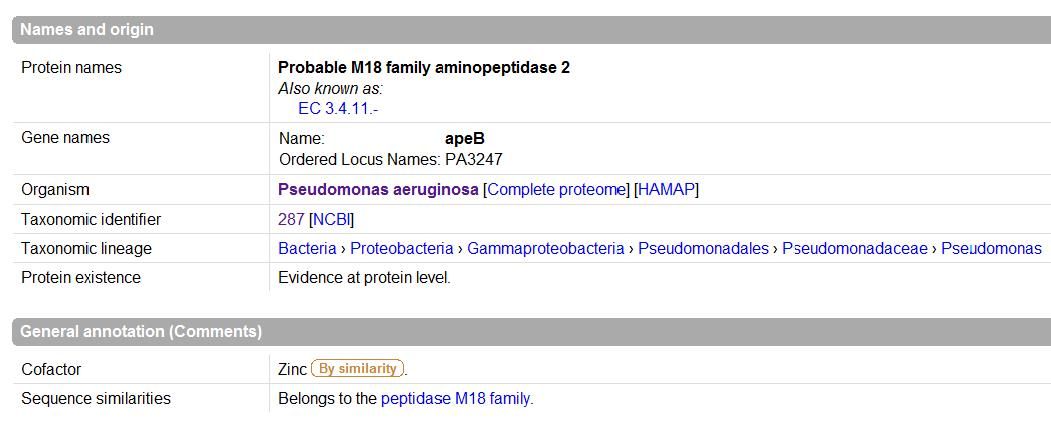
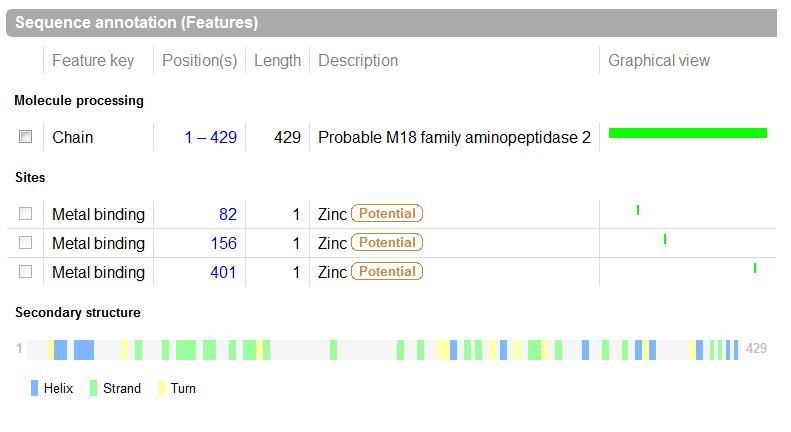
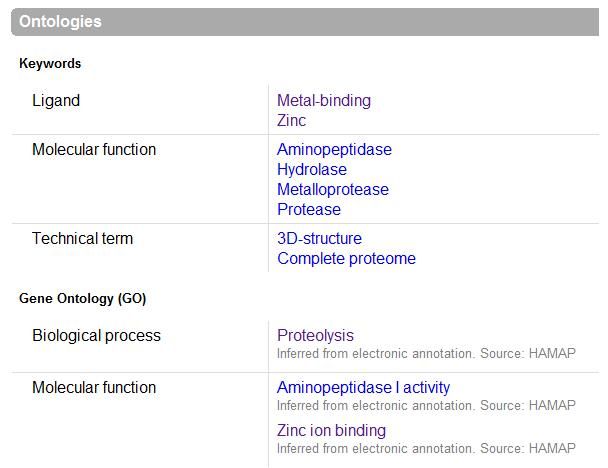
 Fig X. This ProSite scan shows a list of possible active sites with a high probablility of occurence. However they do not seem relevant as they do not take into account the nature of the protein as a metallopeptidase and very few of the predicted catalytic residues feature in this output.
Fig X. This ProSite scan shows a list of possible active sites with a high probablility of occurence. However they do not seem relevant as they do not take into account the nature of the protein as a metallopeptidase and very few of the predicted catalytic residues feature in this output.