DHRS1 Discussion
Sequence
The Short chain dehydrogenase family has evolved over a long period of time and has two main forms, classical SDR and extended SDR. Classic SDR is primarily found in bacteria and is approximately 250 amino acids long and appears to have evolved first, extended SDR's are typically 350 amino acids long and have evolved later. The sequence identity for this family is low, approximately 15 to 20%, This is due to two main reasons, firstly the relatively low number of highly conserved sequences, for example only 4 regions, totaling 8 residues are involved in the ATP binding site, and even within these regions there is some variation of the specific residue, although it is heavily restrained to within the certain groups of amino acids with similar properties and is still almost always conserved to the same residue in these important sites. These sites are infrequent and most of the residues are variable without any significant change in structure or function. Secondly this is partially due to the age of the family and has lead to many point mutations, which have been incorporated over time when the mutation is not fatal to the organism.
One of the mutations incorporated over time is for a subgroup of the family including land based higher organisms such as humans, monkeys, dogs, cats and cows. This is identified by an inserted motif K-[A,S]-F-W-E-x-P-A-S at location 138 - 146. This region does not exist in the classic SDR family and shows variation in the extended family, although is completely conserved within organisms such as those named earlier. The extended SDR family includes some proteobacteria and all the sequences of the anamalia kingdom with the exception of Aedes aegypti. This subgroup suggests that the extended SDR family evolved from the classic SDR family and has continued to evolve. It also suggests that this family was incorporated into higher organisms after this evolution to the extended SDR.
Aedes aegypti was the only eukaryote which incorporated the classic SDR protein in our phylogenetic tree. It is possible that this is due to this particular organisms parasitic nature and this may have increased the oppertunity for lateral gene transfer to occur (Sherwood, 2007). It is possible that this organism did not have the ability to process glucose prior to incorporating this gene from a transfer event and due to this event had a significant evolutionary advantage, causing this protein to be conserved in the organism after transfer.
Structure
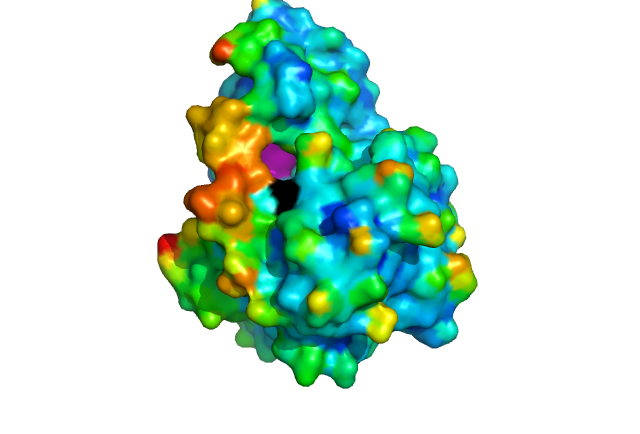
DHRS1 has been crystallized to a resolution of 1.8 Å with an R-value of 0.159. This R-value however was achieved without recording a highly mobile region of 22 amino acids (Fig 5) which covers over the NADP binding site, and may act as a cap protecting the NADP binding site when the enzyme is not in use. This flexible region also covers part of the [G-x(3)-G-x-G] motif.
The SDR family have highly conserved structural features in particular the NADP binding site, central β-strand and [G-x(3)-G-x-G] motif. HSDR1 is representative of the family with all of the required structural elements. It has the usual S-Y-K catalytic triad which is essential for function, binding to NADP (fig 10). It is located in a deep hydrophobic pocket which would help to attract NADP and expel water. The depth of the pocket would be reduced when the mobile part of the protein was open. The central β-strand and [G-x(3)-G-x-G] motif are important for folding of the protein. The [G-x(3)-G-x-G] motif is characteristic of a co-enzyme binding site. NADP a common co-enzyme is already bound at the S-Y-K motif. So the [G-x(3)-G-x-G] motif probably binds something else, it is located on the surface of the protein on the other side of the protein to the S-Y-K catalytic triad. There is a channel in the protein through which the [G-x(3)-G-x-G] motif and the S-Y-K catalytic triad can be aligned (Fig 11). This Channel may open up further when the flexible part of the protein is in a different conformation. This leads us to propose this as a possible substrate binding site although further experiments such site directed mutagenasis and kinetic characterization with possible substrates would be required to validate this hypothesis.
Structural alignments of HSDR1 with other members of the SDR family showed that the most closely structurally related proteins were all involved in reducing substrates (reductases). In particular Glucose reductases appeared frequently (Table 1). All of the proteins that had high structural similarity existed as dimers, tetramers or octamers, indicating that DHRS1 also exists as more than a monomer biologically. Most of the proteins in the SDR family act in tandem with co-enzymes and a highly conserved motif [G-x(3)-G-x-G] that is thought to be a co-enzyme binding site (Wu Q, et al 2001) is also present in DHRS1.
Function
To date, functional studies of DHRS1 have been limited and inconclusive. However, appropriate hypothesises can be formulated by comparing both structural and evolutionary similarities between DHRS1 and other similar known proteins. Also, If similar proteins contain similar structural properties then it can be suggested that DHRS1 shares its function or at least has a similar function to these other proteins.
DHRS1 is a member of the SDR family, which although shows low sequence homology, still has conserved domains. SDR family members can be sub-categorised into sub-families from motif similarities. Classical, extended, intermediate, divergent and complex, are all defined by a sequence motif(s). Studies relating to other similar sub-family proteins offer insight into roles of DHRS1. Classical SDR family members share the following: [ILV]-T-G-x(3)-[AG]-[FILV]-G-x(3)-[AS]-x(2)-[FILMV]-x(3)-G-x(2)-[ILMV] which is found from V12 to L25 in DHRS1. (Kallberg et al. 2002b) Classical SDR also have between 240-280 amimo acids. This suggest that DHRS1 is a classical SDR.
The most important conserved motif is the NAD/NADP (Nicotinamide adenine dinucleotide (phosphate)) binding site. This, almost by itself reserves its function. (See Figure 12). NAD is a coenzyme that transfers the hydride (H-) ion from one group to another. In turn this acts upon the substrate which can chemically change it and is a step in a metabolic pathway. NAD and NADP have roles in metabolism, whether it is fatty acid metabolism, energy metabolism or reductive biosynthesis. (Ying, W. 2008)
By comparing functions of other similar proteins, a hypothesis as to the role of DHRS1 can be made. Using the results from the DALI search, which compares protein structure, the best results were used. (See table 1) 3-oxoacyl-(acyl-carrier-protein) reductase, like DHRS1 is also a member of the SDR protein family and is involved in fatty acid biosynthesis. It also has the conserved binding sequence S-Y-K, in its Rossman fold, which is a structural binding motif and is partly defined by the motif G-x(3)-G-x-G. Also, it has to have three or more beta chains lined by alpha helices. It has a role in biological processes because it binds with coenzyme NAD and catalyses the oxidation of D-glucose to D-beta-gluconolactone which is a step in the glucose metabolic pathway. (Zaccai N et al. 2008)
Structurally, the proteins are also similar (See Figure 13) and a blast search puts its e-value at 9e-023. 3-oxoacyl-(acyl-carrier-protein) reductase has been shown through structural studies to be a tetramer.
Another similar protein, according to the DALI search, is gluconate 5-dehydrogenase which yet again has a similar structural motif in S-Y-K which is the NAD binding domain. It is also contained in a Rossman fold partially define by G-x(3)-G-x-G. Its role is in metabolism is like DHRS1 in that it is an oxidoreductase. (See Figure 14 and 15) | (Merfort M et al. 2006)
Therefore, the possible role of DHRS1, could be as an enzyme that binds a coenzyme NAD or NADP in its binding cleft which has conserved residues over all SDR members. It then binds a substrate which could possibly be glucose. Most of the similar proteins all bind glucose (see table 1) however SDR members are known to bind a number of different substrates from steroids, alcohols, sugars, to aromatic compounds so further experimentation needs to be done before it can be properly known. (Kallberg et al 2002a) DHRS1 works in conjunction with NAD, which donates its hydride ion oxides the substrate into a altered chemical compound. This is important in many biological processes like fat storage/breakdown and metabolism.
Abstract | Introduction | Results | Discussion |
Conclusion | Method | References
Back to Main Page