Discussion - NANP
Discussion
Multiple Sequence Alignment
From the MSA obtained, the organisms with the large gap insertions were isolated to be mainly Bacillus, with the exception
of Symbiobacterium thermophilum. Symbiobacterium is an uncultivable thermophile isolated from compost. Its survival is based mainly on
microbial commensalisms 5. This bacterium can only grow in vitro, if it is co-cultured with Bacillus species
bacteria 5. This could therefore explain its genetic association with Bacillus, as observed from the sequence
alignment. However, interestingly, Bacillus is classified as Gram-positive, while Symbiobacterium is a Gram-negative bacterium. As
observed from the sequence alignment, other Gram-negative bacterium protein sequences (Vibrio species) do not contain the large gap
insertion at the 91st to 114th amino acid positions, with the exception of Symbiobacterium. Hence, more genetic (and
even functional) analysis might be necessary to determine the hydrolase protein relationship between the Gram-positive Bacillus with the
Gram-negative Symbiobacterium.
Phylogenetic Tree
From the Rectangular Cladogram view of the tree, it was observed that there were two main Domains — Procaryotes and Eucaryotes. This would also
be the root and first branching point of the phylogenetic tree.
The invertebrates (of Phylum Arthropoda) would be the first branching point for the eucaryotes in this tree.
From there, further branching occurs into the vertebrates (of Phylum Chordata). This would then be further branched into Osteichthyes (bony
fish) and Tetrapoda (four-limbed vertebrates) Superclasses.
For the prokaryotic domain, mainly branching occurs between Gram-positive (Bacillus spp.) and Gram-negative (Vibrio spp.) bacteria.
Hence, it can be generally deduced that the Neu5Ac (hydrolase) protein is non-evolutionary specific, as it is observed to be present in almost
all main Phyla and Classes of organisms from the two main Procaryotic and Eucaryotic Domains. Its functional significance would therefore be a
general one.
Bootstrapping
Tree bootstrapping is necessary to test for the reliability of the branching patterns and distances formed on the phylogenetic tree. This was
done by making "pseudoreplicates" of multiple sequence alignments of up to 100 sets. The distance matrices were recalculated using these d
duplicate alignment values to generate a bootstrap tree, which can be used to compare the branching patterns and distances with the original
phylogenetic tree.The bootstrap values (in percentage) obtained on each branch, signify branching confidence. Bootstrap values of 95% equate to
full branching confidence; 75% value equates to 95% branching confidence; 60% value equates to much lowered branching confidence; while 50%
value would render no branching confidence.
Functional Analysis
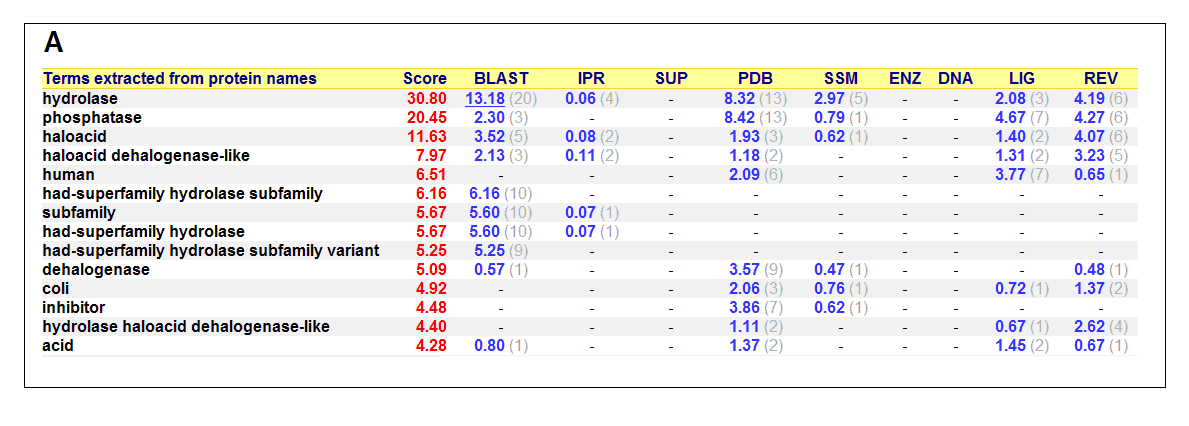
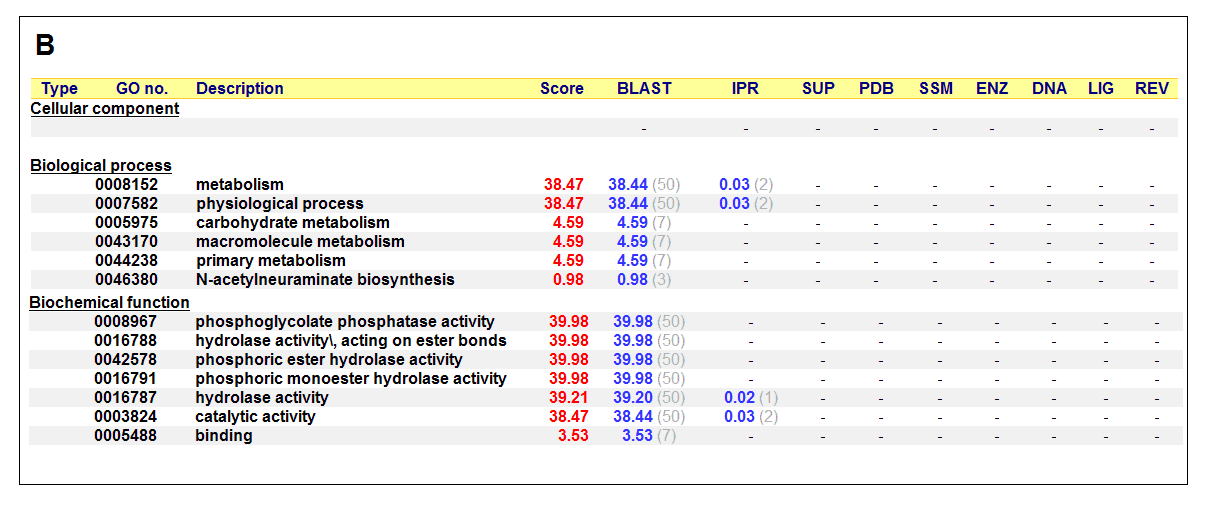 Figure 17. (A) List of all matched protein name terms for 2gfh. (B) List of all matched Gene Ontology terms for 2gfh. The score in
Figure 17. (A) List of all matched protein name terms for 2gfh. (B) List of all matched Gene Ontology terms for 2gfh. The score in
red is a measure of how strongly the term is predicted from the hits obtained by the different methods. The scores in blue show each
method’s contribution to the total score (with the number of relevant sequences/structures shown in brackets in grey).
(http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/)
The predicted function based on the evolution and structure, illustrate that 2gfh is a hydrolase. Profunc searches (Figure 17) on 2gfh also
show that it possesses hydrolase activity. The highest score for Gene Ontology (Figure 17) states it used for metabolism and possesses
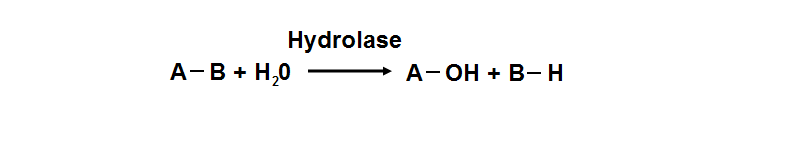
phosphoglycolate phosphatase activity. Hydrolyase is an enzyme which catalyzes hydrolysis reaction (Figure 18), which is the addition of the
hydrogen and hydroxyl ions of water to a molecule with its consequent splitting into two or more simpler molecules. Hydrolase is the systematic
name for any enzyme of EC class 3.
Figure 18. Hydrolyase catalyze the hydrosis of the chemical bond between A and B, resulting of 2 simple molecules.
Neu5Ac phosphatase belongs to the HAD family, HAD is a vast superfamily of largely uncharacterized enzymes, with a few members shown to possess
phosphatase, phosphoglucomutase, phosphonatase, and dehalogenase activities 6. HAD-like hydrolases represent the largest family of
predicted small molecule phosphatases encoded in the genomes of bacteria, archaea, and eukaryotes, with 6,805 proteins in data bases 7.
HADs share little overall sequence similarity (15–30% identity), but they can be identified by the presence of three short
conserved sequence motifs 7. Most of the characterized HADs have phosphatase activity (CO–P bond hydrolysis), catalyze dehalogenase
activity (C–halogen bond hydrolysis), phosphonatase (C–P bond hydrolysis), and phosphoglucomutase (CO–P bond hydrolysis and intramolecular
phosphoryl transfer) reactions 6.
In the study conducted by Maliekal et al (Figure 19), they compared the alignment of the first 280 amino acids of rat and human Neu5Ac-9-P
phostphatase with other 2 homologous sequences.
Figure 19. Alignment of rat and human Neu5Ac-9-P phosphatase with homologous sequences. The following sequences are aligned: Rattus
norvegicus (Rnor, gi-34859431), Homo sapiens (Hsap, gi-23308749), Xenopus laevis (Xlae, gi-46250196), Danio rerio (Drer, gi-
63101958), and Drosophila melanogaster (Dmel, gi-28381565). Only the first 280 residues of the latter sequence are shown. Completely
conserved residues are shown in boldface type. Asterisks indicate the extremely conserved residues in phosphatases of the HAD family 8.
The MSA done by Maliekal et al shows that the Neu5Ac-9-Pase orthologs shared the three motifs found in phosphatases of the HAD family,
namely a 1st motif comprising two extremely conserved aspartates (D), a 2nd motif comprising a conserved serine (S) or
threonine (T), and a 3rd motif comprising a conserved lysine (K) and two conserved aspartates (D) 8. The first aspartate
in the first motif forms a phosphoaspartate during the catalytic cycle 9. These findings suggested therefore that the HDHD4 protein
was a phosphatase. The first aspartate in the first motif forms a phosphoaspartate during the catalytic cycle 10. In our MSA (Figure
16), the several conserved motifs that shared great similarity to the study done by Maliekal et al. These findings suggested therefore that
Neu5Ac-9-P phosphatase protein is a phosphatase
Phosphatases of the HAD family are dependent on the presence of Mg2+ and Ca2+ inhibits
their activity by replacing Mg2+ and preventing the nucleophilic attack by the aspartate that covalently binds the
phosphate group 8. Phosphatases that form a phosphoenzyme during the catalytic cycle, are inhibited by vanadate 11.
Vanadate (VO43−), formed when V2O5 is dissolved in water at alkaline pH, appears to inhibit enzymes
that process phosphate.
The presence of a protein sharing at least about 50% sequence identity with rat or human Neu5Ac-9-P phosphatase in the genomes of mammals,
chicken, xenopus, and fishes indicates that sialic acid synthesis proceeds via the 9-phosphate intermediate in these species 8. This
is consistent with the finding that the genome of vertebrates comprises a gene encoding the bifunctional enzyme UDP- N-acetylglucosamine-2-
epimerase or N-acetylmannosamine kinase 8.
In bacteria, E. coli genome encodes five membrane-bound and 23 soluble HAD-like hydrolases, representing about 40% of the E. coli
proteins with known or predicted small molecule phosphatase activity 12. The metabo lites hydrolyzed by HADs are intermediates of
various metabolic pathways and reactions (glycolysis, pentose phosphate pathway, gluconeogenesis, and intermediary sugar and nucleotide
metabolism).
E. coli HADs hydrolyze a wide range of phosphorylated metabolites, including carbohydrates, nucleotides, organic acids, and coenzymes.
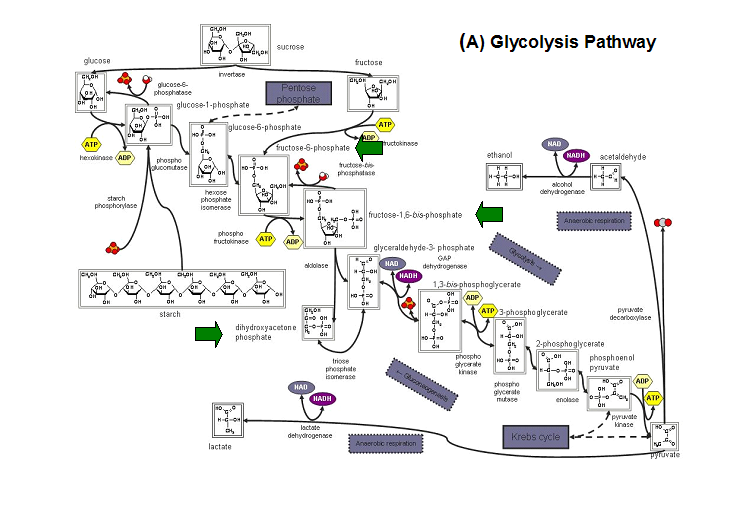
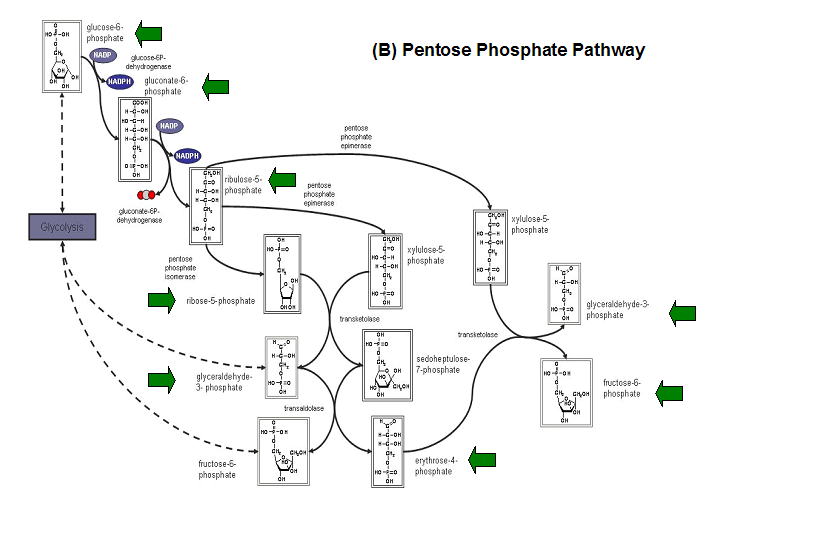
Studies have shown that the most common substrates in metabolism such as glycolysis and pentose phosphate pathway (Figure 18). These enzymes
were fructose-1-phosphate, glucose-6-phosphate, mannose-6-phosphate, 2-deoxyglucose-6-phosphate, fructose-6- phosphate, ribose-5-phosphate, and
erythrose- 4-phosphate 13.
Figure 20. The schematic diagrams of glycolysis and pentose phosphate metabolic pathways. The green arrows show the substrates that are hydrolyzed by HADs (A) Glycolysis pathway with substrates that are hydrolyze by HADs: glucose 6-phosphate, fructose 6-phosphate and dihydroxyacetone phosphate. (B) Pentose phostphate pathway with substrates that are hydrolyze by HADs: glucose-6-phosphate, fructose-6-phosphate, dihydroxyacetone phosphate, glyceraldehyde-3-phosphate, gluconate 6-phosphate and erythrose-4-phosphate.