Evolutionary analysis
Whole sequence alignment
Score E Sequences producing significant alignments: (bits) Value gb|AAH58177.1| Haloacid dehalogenase-like hydrolase domain conta... 462 e-128 ref|NP_084102.1| haloacid dehalogenase-like hydrolase domain con... 462 e-128 dbj|BAE28142.1| unnamed protein product [Mus musculus] 460 e-128 pdb|2HO4|A Chain A, Crystal Structure Of Protein From Mouse Mm.2... 453 e-126 ref|NP_001014173.1| hypothetical protein LOC361351 [Rattus norve... 446 e-124 ref|XP_537270.1| PREDICTED: similar to haloacid dehalogenase-lik... 437 e-121 ref|NP_001030194.1| haloacid dehalogenase-like hydrolase domain ... 426 e-118 ref|XP_001148863.1| PREDICTED: haloacid dehalogenase-like hydrol... 424 e-117 emb|CAH93402.1| hypothetical protein [Pongo pygmaeus] 424 e-117 ref|NP_115500.1| haloacid dehalogenase-like hydrolase domain con... 421 e-116 ref|XP_001086150.1| PREDICTED: similar to haloacid dehalogenase-... 418 e-115 ref|XP_001086254.1| PREDICTED: similar to haloacid dehalogenase-... 413 e-114 dbj|BAB22395.1| unnamed protein product [Mus musculus] 410 e-113 ref|XP_414700.2| PREDICTED: hypothetical protein [Gallus gallus] 390 e-107 gb|AAS66213.1| LRRG00122 [Rattus norvegicus] 389 e-107 ref|NP_001013491.1| hypothetical protein LOC541346 [Danio rerio]... 371 e-101
gi:14149777 is human version.
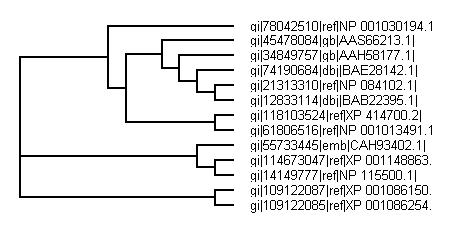
Phylogenetic tree from CLUSTALX
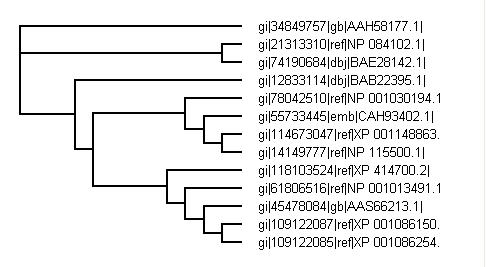
Phylogenetic tree from PHYLIP
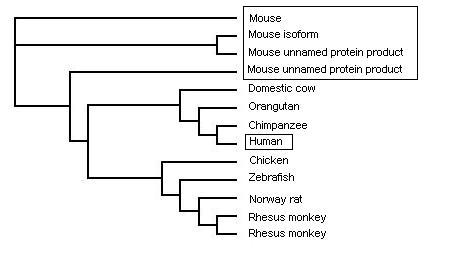
Phylogenetic tree from PHYLIP - by species
BLAST for first 100 nucleotides
Score E Sequences producing significant alignments: (Bits) Value gi|114794442|pdb|2HO4|A Chain A, Crystal Structure Of Protein... 175 8e-43 gi|34849757|gb|AAH58177.1| Haloacid dehalogenase-like hydrola... 174 2e-42 gi|21313310|ref|NP_084102.1| haloacid dehalogenase-like hydro... 172 4e-42 gi|74190684|dbj|BAE28142.1| unnamed protein product [Mus musculu 172 4e-42 gi|78042510|ref|NP_001030194.1| haloacid dehalogenase-like hy... 167 1e-40 gi|57089151|ref|XP_537270.1| PREDICTED: similar to haloacid d... 167 1e-40 gi|12833114|dbj|BAB22395.1| unnamed protein product [Mus musculu 167 2e-40 gi|75040931|sp|Q5R4B4|HDHD2_PONPY Haloacid dehalogenase-like ... 162 4e-39 gi|62079029|ref|NP_001014173.1| hypothetical protein LOC36135... 162 7e-39 gi|114673047|ref|XP_001148863.1| PREDICTED: haloacid dehaloge... 161 9e-39 gi|85861235|ref|NP_001034291.1| haloacid dehalogenase-like hy... 159 4e-38 gi|14149777|ref|NP_115500.1| haloacid dehalogenase-like hydro... 159 6e-38 gi|114673057|ref|XP_001148387.1| PREDICTED: haloacid dehaloge... 158 9e-38 gi|109122087|ref|XP_001086150.1| PREDICTED: similar to haloac... 155 8e-37 gi|109092050|ref|XP_001088800.1| PREDICTED: similar to haloac... 154 1e-36
BLAST search for whole sequence turns out a high number of similar matches. Top few matches were used to generate a phylogenetic tree.
Both phylogenetic trees from CLUSTALX and PHYLIP both show similar results with PHYLIP being known as the more accurate version for study.
It is not suprising that the closest related to the human is the chimpanzee's gene and orangutan. It has long been known that these are the two closest related animals to humans and so this result is to be expected. The suprising result is for the mouse genes. There are 4 genes all closely related and branching off with 3 being off the same branch with slight differences. All the mouse genes are similar with the top one a confirmed and studied example and subsequent genes all showing very close matches. With the high degree of nucleotide match and studied and identified genes of many species it is arguable that the genes labeled as "predicted" and "hypothetical" are of that function. The mouse gene seems to be the genes closest to the ancestorial gene with the identified and yet studied forms.
BLAST search for first 100 nucleotides is run to determine the degree of conservance between strains. Values were much lower indicating less matches but the matches were still very much the same types of proteins. This suggests that the first 30 odd amino acids do vary a bit between different species.