Evolutionary analysis of 2ece
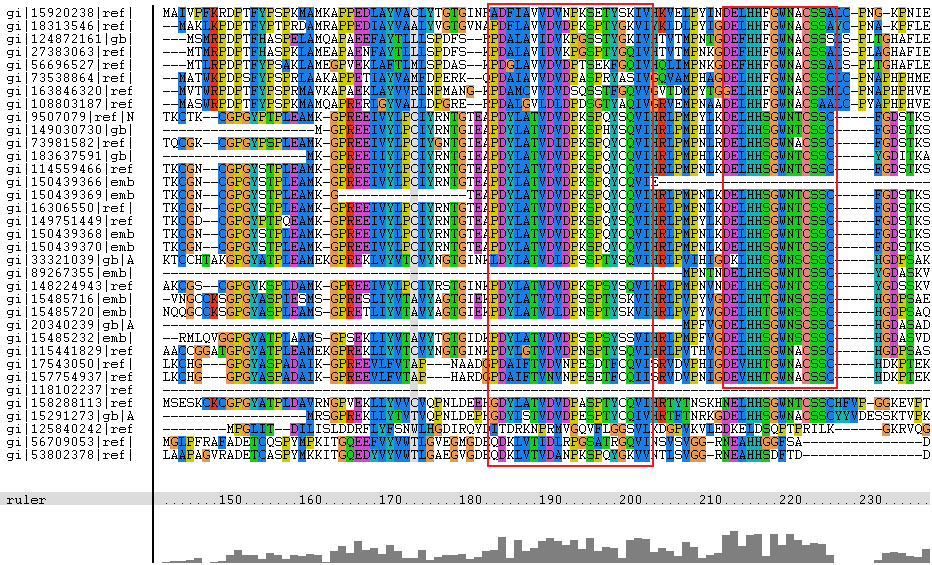
The amino acid sequence of selenium binding protein 2ece, sourced from the organism Sulfolobus tokodaii str. 7, was compared with other proteins using a psi blast(Media:psiblast.txt), with hits from the results hand selected (to show biological diversity) to go through a multiple sequence alignment in clustalx with portions of the alignment shown below
Figure 3.1 ClustalX msa of selected proteins against selenium binding protein
The segments in red boxes are reasonably highly conserved amongst the organism, and the segment in a brown box apperas to be a common insertion shared between some organisms. The protein sequences are labelled with gi numbers, the table below associates organisms to specific gi numbers.
Table 3.1 Association between gi numbers and organism species
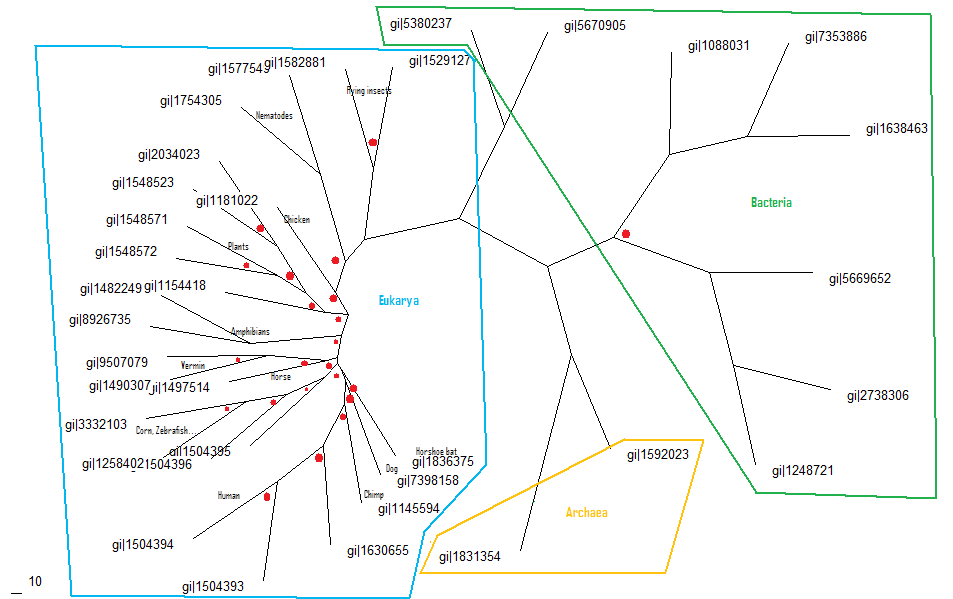
The shown alignment was prepared into a phylogenic tree, shown below, using the nieghbourhood joining method, then creating a consensus tree with bootstrap values. The three kingdoms are circled and labelled and nodes with bootstrap values of less than 75% shown with red dots.
Figure 3.2 Phylogenic Tree
The consense data can be found here Media:consense.txt
As shown in the phylogenic tree selenium binding protein is widespread throughout the three domains, it seems a bias of eukarya was chosen for the analysis though this doesn't truly represent the diversity of the protein. Most organsims appear to be where they should inthe phylogenic tree which suggests vertical gene transfer though some species are in unexpected places suggesting some lateral gene transfer took place. This can be seen with corn, zebrafish and a couple of human recorded proteins ending up on the same branch, and a chicken protein ending up near the plant kingdom. Also across kingdoms some results were unexpected such as the nematodes were expected to be closer to the bacteria domain than the flying insects, and the plant kingdom seperating the animalia kingdom. This may be misinterperated as lateral gene transfer (across ancestors) but can easily be explained by the limited capabilities of the bioinformatics programs used. Though there may be evidence of lateral gene transfer with particular expression of the gene in symbiosis between plants and bacteria (Agalou, 2005)
As seen on the phylogenic tree it has a fair few boot strap values lower than 75% with most of them in the eukarya domain. This can be explain by the selection of proteins used, and that the multiple sequence alignment may not have made the best alignment possible. These low values explain the oddities of the tree, which are mainly prevalent in the eukarya domain. Though overall most of the phylogenic tree is believable, even though the confidence of the tree is low.
Back to resultsResults 2ece
Back to menuSelenium binding protein