Fascin Evolution: Difference between revisions
No edit summary |
|||
| (11 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
Blast Results show that fascin 1 is very highly conserved throughout the Eukarya Domain providing 112 hits all with excellent E value and High scores | Blast Results show that fascin 1 is very highly conserved throughout the Eukarya Domain providing 112 hits all with excellent E value and High scores | ||
__TOC__ | |||
[[Image:Flascin_1.jpg]] | [[Image:Flascin_1.jpg]] | ||
Sample Fascin sequence (Text File) | |||
[[Media:fascin1_fasta.txt]] | |||
FASTA sequences from BLAST (Text File) | |||
[[Media:FASTA_fascin1_all.txt]] | |||
== Multiple Alignment == | == Multiple Alignment == | ||
| Line 14: | Line 22: | ||
== Phylogency Trees == | == Phylogency Trees == | ||
Before any deletion of sequences occured, A tree was formed using 100-replications Dayhoff neighbor-joining method as a base line. | |||
[[Image:Facscin_broad_bootstrap.jpg]] | |||
| Line 23: | Line 42: | ||
Neighbor-method with Bootstrap values, compressed. Sequences with below 50 were deleted. | Neighbor-method with Bootstrap values, compressed. Sequences with below 50 were deleted. | ||
[[Image: | [[Image:Fascin_bootstrap_animaliainsecta.jpg]] | ||
[[Fascin Evolution | Evolution]] | [[Fascin Function | Function]] | [[Fascin Structure | Structure]] | |||
<br>[[Fascin 1 | Back to main page]] | |||
Latest revision as of 02:03, 16 June 2009
BLAST
Blast Results show that fascin 1 is very highly conserved throughout the Eukarya Domain providing 112 hits all with excellent E value and High scores
Sample Fascin sequence (Text File)
FASTA sequences from BLAST (Text File)
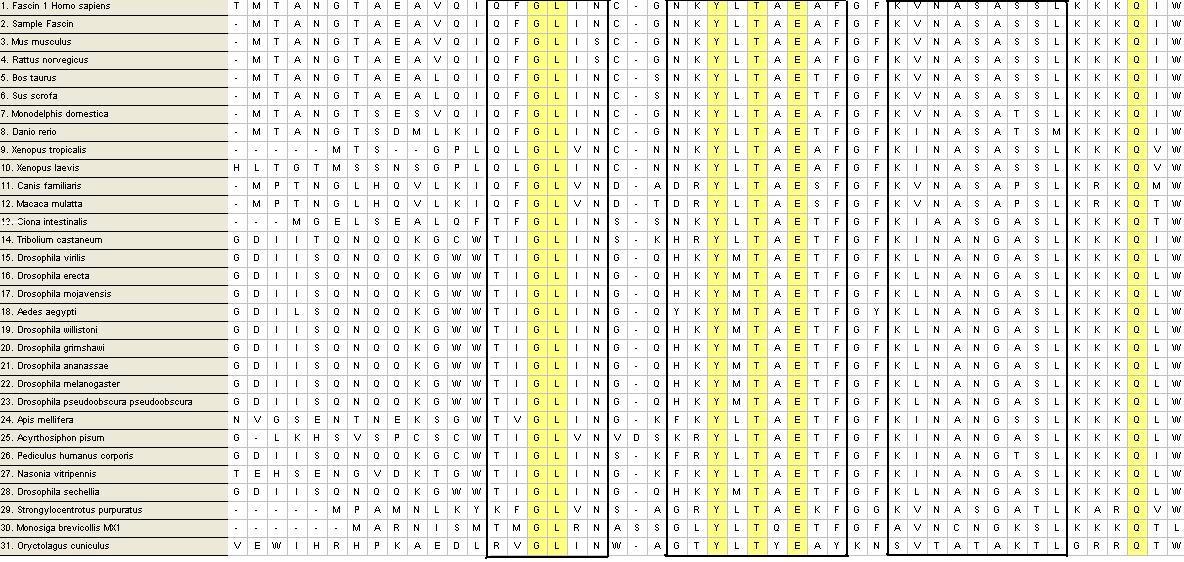
Multiple Alignment
From a possible 100 FASTA sequences, were cut down to 31. The process of elimination was conducted by the deletion of sequences with too many bp before and after the sample sequence, followed by the deletion of sequences with large segments of gaps in conserved and variable regions of the sequence alignment. Fascin domains were found via a protDOM search and compared to the remaining sequences.
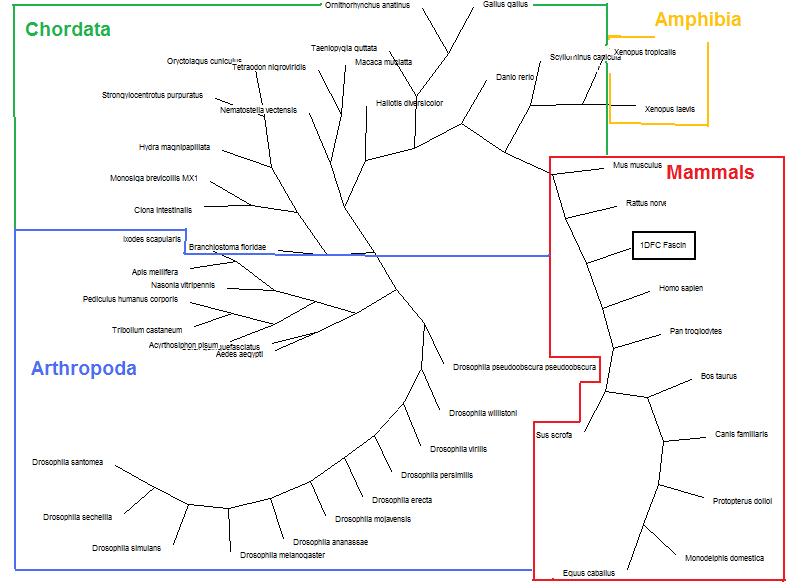
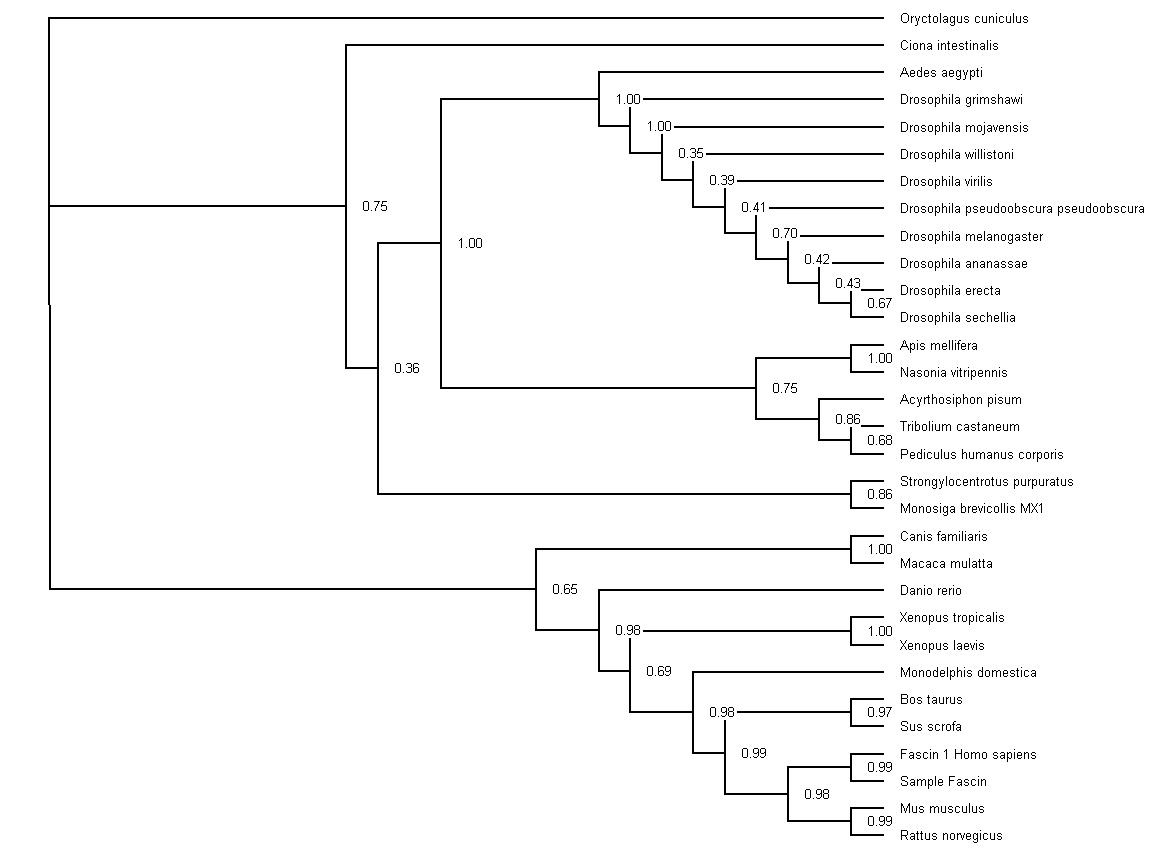
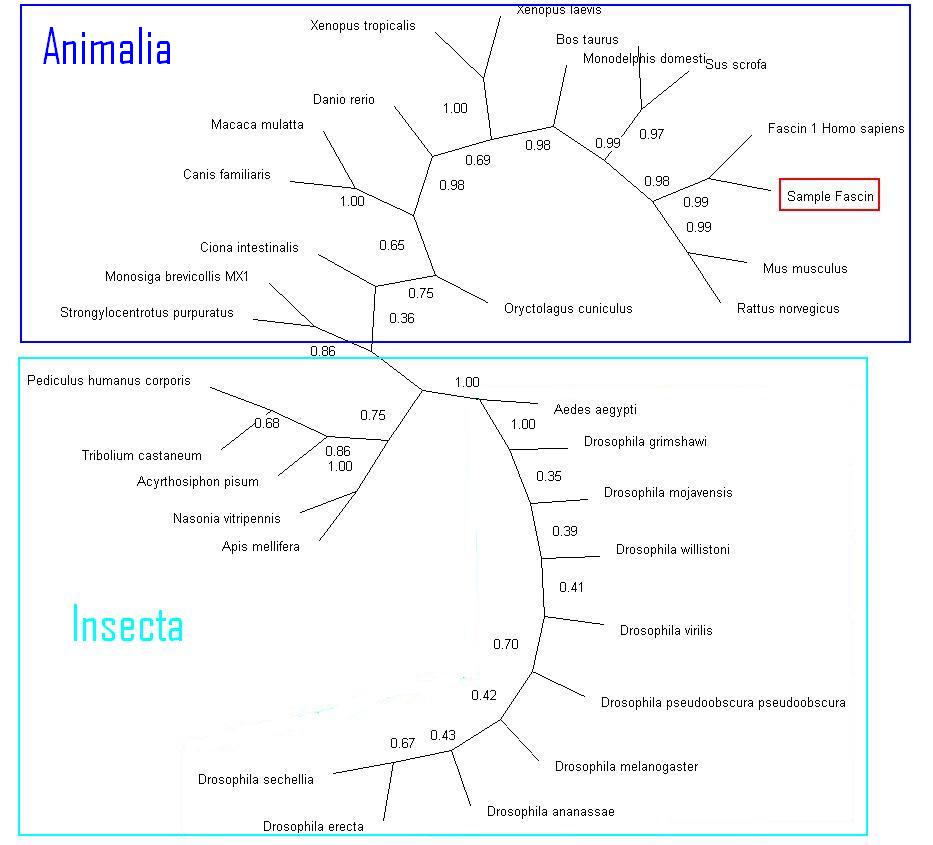
Phylogency Trees
Before any deletion of sequences occured, A tree was formed using 100-replications Dayhoff neighbor-joining method as a base line.
Neighbor-method with Bootstrap values (1 = 100) Results were run at 500 replications
Neighbor-method with Bootstrap values, compressed. Sequences with below 50 were deleted.