Hypothetical protein Introduction
The sugars fucose and ribose both play an important role in the function of cells. Ribose is used in a range of cells for differing purposes, such as DNA, RNA synthesis and limited nutritional purposes. Fucose on the other hand plays an important role in cell receptors involved with cell to cell adhesion. Both of these sugars require transport systems to gain access into the cell. These transport systems used are called ATP-cassette binding (ABC) systems. These ATP-binding cassette (ABC)-type systems have been found to mediate the uptake of solutes such as amino acids, peptides, ions, vitamins, and sugars in a wide range of bacteria (Tuner et al. 1999).These ABC transport systems are found in all forms of life, both eukaryotes and prokaryotes. The structure of a typical prokaryote type ABC transporter consists of three types of components: two peripheral proteins that bind and hydrolyze ATP, two integral proteins each having six transmembrane segments and a periplasmic substrate binding protein (Igarashi et al 2004).
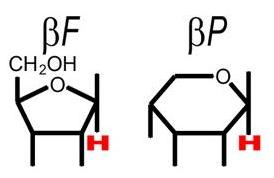
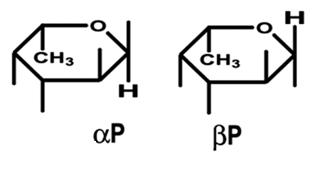
The FucU/RbsD superfamily of proteins are involved with the transport of fucose and ribose into the cell. These proteins are thought to be involved with the anomeric conversion of these two sugars . RbsD proteins convert beta-purine ribose into its furine counterpart (see figure 1) which is needed for the next step in metabolism. FucU proteins on the other hand catalyze the change of alpha-purine fucose into the beta form (see figure 2). The beta form is needed for use in the salvage pathway to form a number of important receptors and secretions (Kim et al. 2003).
Figure 1.This shows the difference between the 5 ring franose and the six ring pruanose form of ribose. (Kim et al. 2003)
Figure 2.This shows the difference between the alpha and beta form of fucose. (Ryu et al. 2007)
The 2ob5A protein (Hypothetical protein LOC282969 isoform 1) is found to be a member of the FucU/RbsD superfamily. The aim of this report is to provide information covering the evolutionary history of the protein through the construction of a phylogenic tree, as well as provide insight into its structure and function through the use of numerous bioinformatics techniques. The information provided will hopefully assist in future research of this protein and related proteins.
Abstract | Introduction | Method |
Results | Discussion | Conclusion | References