Kenn
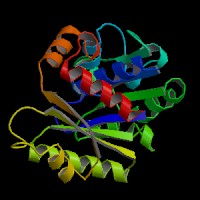
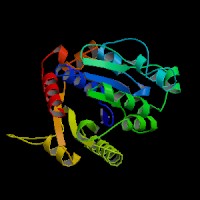
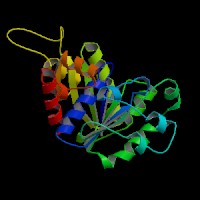
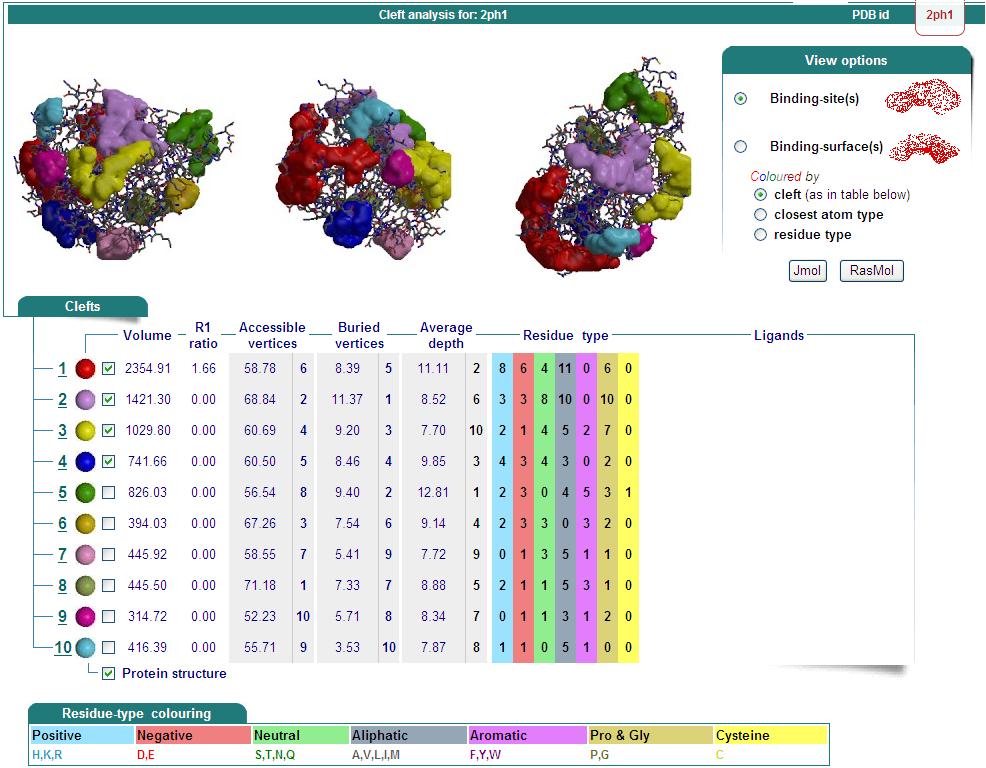
Structure Summary
NESG ID: GR165
PDB ID: 2ph1
Deposition date: 2007-04-10
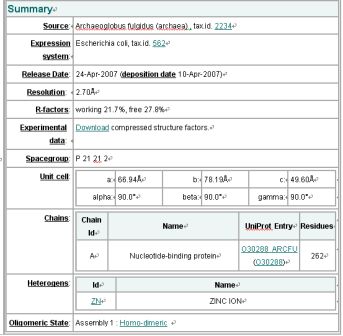
Common Name: Nucleotide-binding protein
Length (a.a.): 240
Organism: Archaeoglobus fulgidus
SwissProt / TrEMBL ID: O30288_ARCFU
Oligomerization: monomer
Molecular weight: 26087
Secondary Structure Elements: alpha helices: 7-14, 32-45, 61-65, 111-122, 143-151, 167-178, 211-217, 230-237 beta strands: 83-84, 91-93, 50-54, 131-135, 20-24, 155-160, 184-189, 222-225, 74-76, 79-81, 194-195, 202-203
Resolution: 2.70 Å R-factor: 0.217 R-free: 0.278
EMI result
DaliLite: Structural Neighbours
Query: mol1A MOLECULE: NUCLEOTIDE-BINDING PROTEIN;
Matches are sorted by Z-score. Similarities with a Z-score lower than 2 are spurious.
Summary
No: Chain Z rmsd lali nres %id Description
1: 2ph1-A 47.3 0.0 247 247 100 MOLECULE: NUCLEOTIDE-BINDING PROTEIN;
2: 1g3q-A 20.0 2.7 207 237 21 MOLECULE: CELL DIVISION INHIBITOR;
3: 1ion-A 19.8 2.6 206 243 20 MOLECULE: PROBABLE CELL DIVISION INHIBITOR MIND;
4: 1hyq-A 19.6 3.1 197 232 25 MOLECULE: CELL DIVISION INHIBITOR (MIND-1);
5: 1g3r-A 19.6 2.7 206 237 20 MOLECULE: CELL DIVISION INHIBITOR;
6: 1cp2-A 19.0 2.8 204 269 16 MOLECULE: NITROGENASE IRON PROTEIN;
7: 1g1m-B 18.9 2.8 202 289 18 MOLECULE: NITROGENASE IRON PROTEIN;
8: 1xd9-A 18.8 2.8 203 289 17 MOLECULE: NITROGENASE IRON PROTEIN 1;
9: 1cp2-B 18.8 2.7 203 269 16 MOLECULE: NITROGENASE IRON PROTEIN;
10: 1nip-B 18.4 2.8 202 287 18 NITROGENASE IRON PROTEIN
11: 1de0-A 18.4 2.8 203 289 18 MOLECULE: NITROGENASE IRON PROTEIN;
12: 1xdb-A 18.3 2.9 204 289 18 MOLECULE: NITROGENASE IRON PROTEIN 1;
13: 1de0-B 18.3 2.8 203 289 18 MOLECULE: NITROGENASE IRON PROTEIN;
14: 2bej-A 18.1 3.0 202 245 26 MOLECULE: SEGREGATION PROTEIN;
15: 1n2c-F 18.1 2.9 203 274 18 MOLECULE: NITROGENASE MOLYBDENUM-IRON PROTEIN;
16: 1n2c-E 18.1 3.0 205 274 18 MOLECULE: NITROGENASE MOLYBDENUM-IRON PROTEIN;
17: 1g5p-B 18.1 2.7 202 289 18 MOLECULE: NITROGENASE IRON PROTEIN;
18: 1g5p-A 18.1 2.8 201 286 17 MOLECULE: NITROGENASE IRON PROTEIN;
19: 1g1m-A 18.1 2.8 202 287 18 MOLECULE: NITROGENASE IRON PROTEIN;
20: 2nip-B 18.0 2.7 201 289 18 MOLECULE: NITROGENASE IRON PROTEIN;
21: 2nip-A 18.0 2.8 201 286 17 MOLECULE: NITROGENASE IRON PROTEIN;
22: 1nip-A 18.0 2.8 203 283 18 NITROGENASE IRON PROTEIN
23: 1n2c-G 18.0 2.9 203 274 18 MOLECULE: NITROGENASE MOLYBDENUM-IRON PROTEIN;
24: 1m34-F 18.0 2.8 201 274 18 MOLECULE: NITROGENASE MOLYBDENUM-IRON PROTEIN ALPHA CHAIN;
DALI output describes the following :
Z score , the statistical significance of the similarity between protein-of-interest and other neighbourhood protines. The program optimises a weighted sum of similarities of intramolecular distances.
Root Mean Square Distance (RMSD), root-mean-square deviation of C-alpha atoms in the least-squares superimposition of the structurally equivalent C-alpha atoms. As in indicated in DALI, rmsd is not optimised and is only reported for information.
lali, the number of structurally equivalent residues.
nres, or the total number of amino acids in the hit protein.
%id - percentage of identical amino acids over structurally equivalent residues.
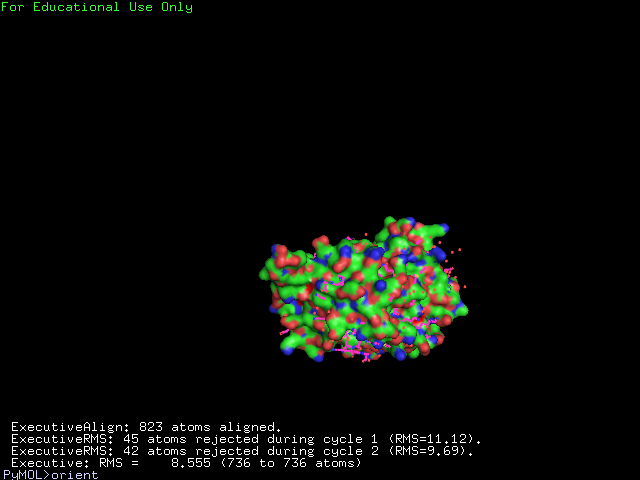
The structure of 1g3q is the most similar to 2ph1 according to the result from Dali.This figure shows the alignment of 2ph1 and 1g3q with RMS 0.55.
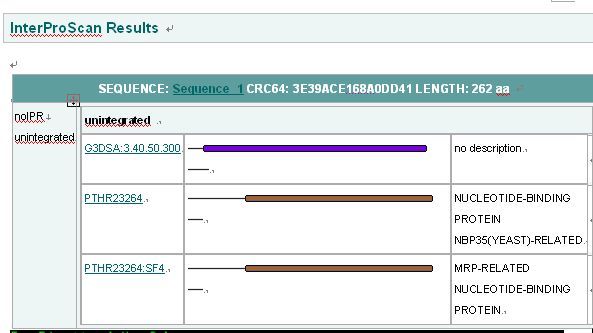
Interpro Result
PUBSUM:
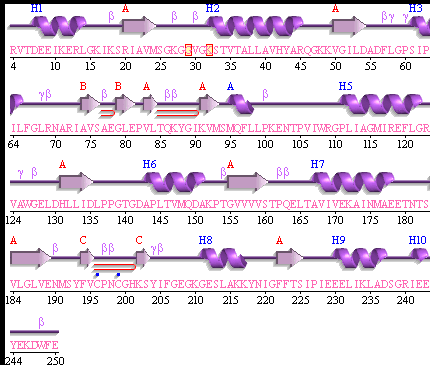
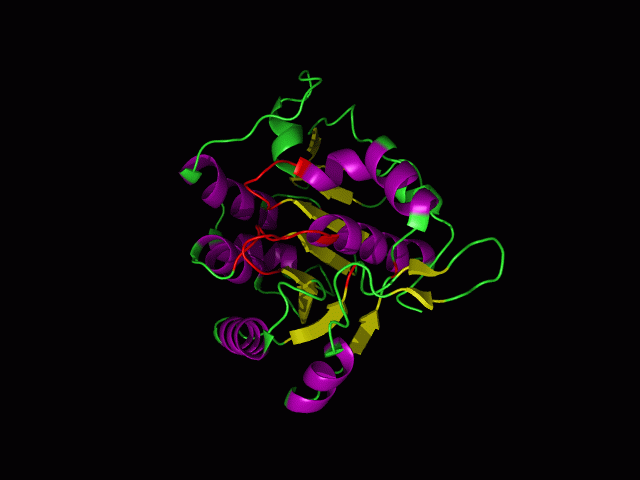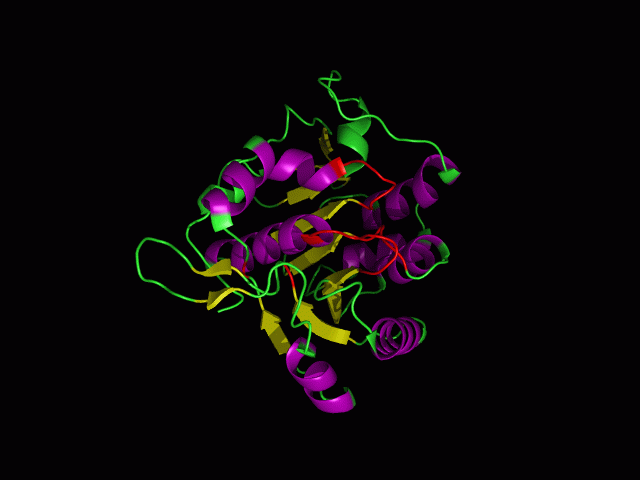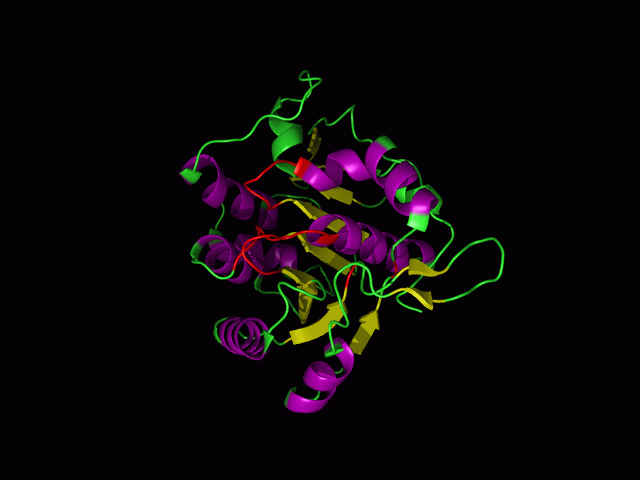
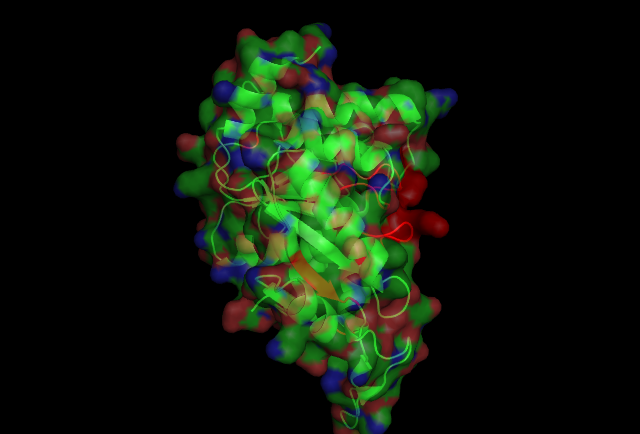
This picture shows the secondary structure of 2ph1, the alpha helixes were shown in purple, and the beta sheets are shown in yellow.
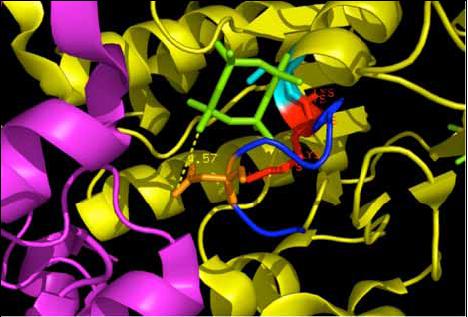
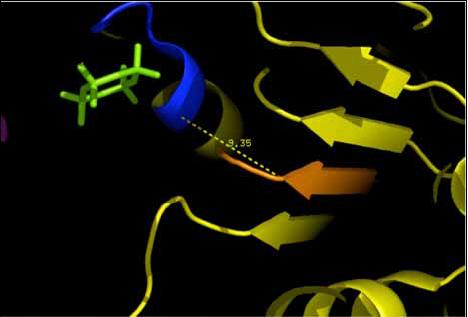
The 2ph1 molecule with ATP binding