LOC56985 Evolution Main
BLASTP Search Results
The BlastP search results were mainly comprised of hypothetical or predicted proteins. The closest protein sequence results that weren’t classified as “hypothetical” was the Norway Rat liver manganese (II)-dependent ADP-ribose/CDP-alcohol pyrophosphatase (e-173), the calcineurin-like phosphoesterase family protein of Arabidopsis thaliana (e-53), a Twin-arginine translocation pathway signal in Mesorhizobium bacteria (e-26) and metallophosphoesterases in Rhizobium leguminosarum (e-23), Methylobacterium species (e-21), Pelodictyon phaeoclathratiforme (e -14) and Chlorobium limicola (e-13).
The initial top 60 blast search results, given in FASTA format, can be found in the link below.
For the purpose of creating a phylogenetic tree, sequences from the same species were removed and any sequences containing large gaps in the intial CLUSTAL alignment were also removed. The edited version of the blast search results used for the final clustal alignment can be seen below. Please note that {H} stands for hypothetical protein and {P} stands for predicted protein.
CLUSTALX Alignment
The results for the CLUSTALX alignment can be viewed here.
"The line above the ruler is used to mark strongly conserved positions. Three characters ('*', ':' and '.') are used:
'*' indicates positions which have a single, fully conserved residue
':' indicates that one of the following 'strong' groups is fully conserved:- STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, FYW
'.' indicates that one of the following 'weaker' groups is fully conserved:- CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, FVLIM, HFY
These are all the positively scoring groups that occur in the Gonnet Pam250 matrix. The strong and weak groups are defined as strong score 0.5 and weak score =<0.5 respectively." (Thompson, J.D. et. al. 1997)
Notice the cluster of residues that are invariable (ie '*') across all species. It is likely that these particular residues are important for the Structure of this protein.
Protein Tree
Multiple alignment results from ClustalX were used to create and bootstrap a tree. Bootstrapping is a means of assessing the confidence or accuracy of the tree (Efron, B. et. al. 1996). 1000 bootstrap trials were performed and the results were viewed on Tree View.
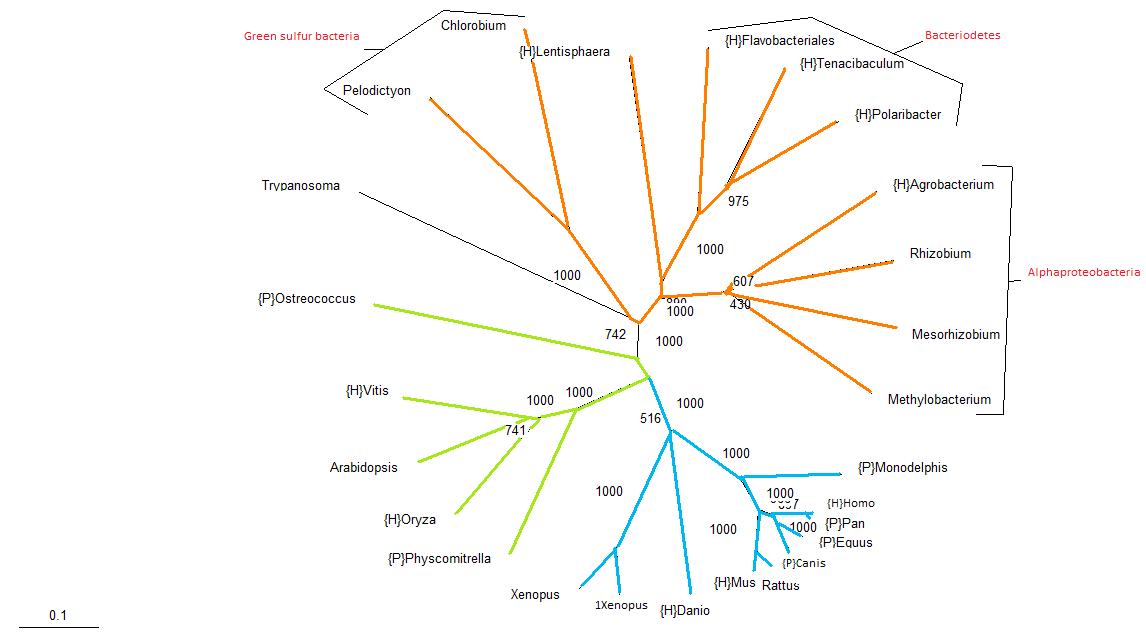
| Protein Tree for LOC56985 |
|---|
In this unrooted tree, bacterial species are shown by orange branches, plants are shown by green branches and vertebrates are shown by blue branches. One can see from this tree that the assortment of organisms is generally congruent with traditional phylogenetic trees, with the exception of the Trypanosoma, which is an eukaryote that has been grouped together with the prokaryotes.
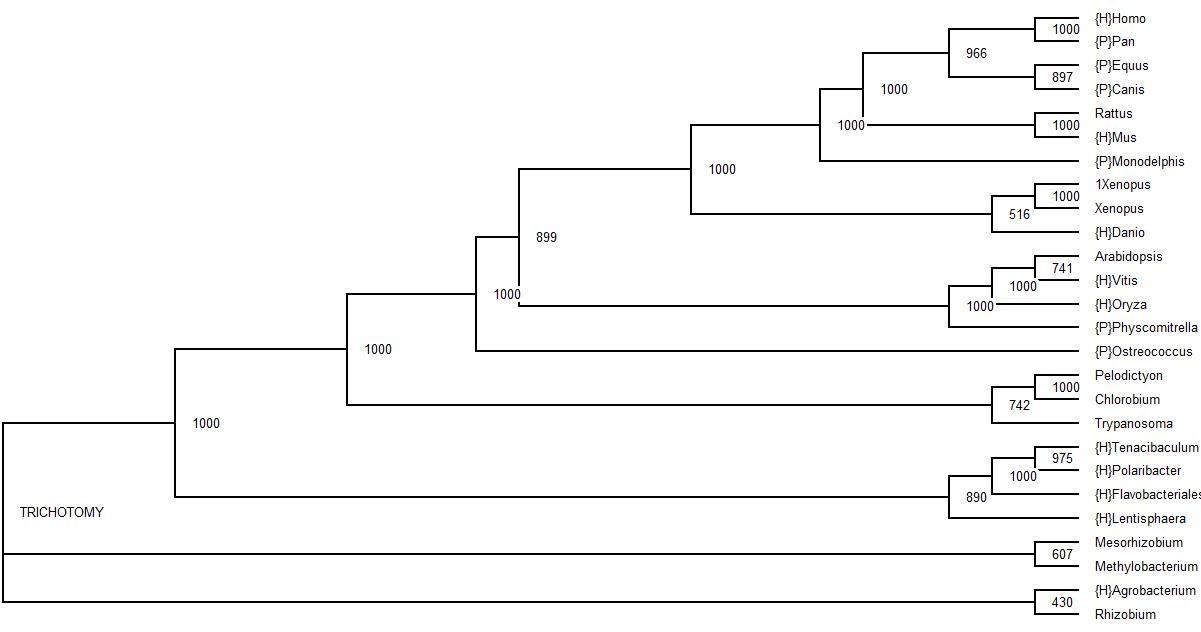
The bootstrap values are also shown, though some overlap is evident, thus the bootstrap values can be more clearly seen in the tree below.
| Protein Tree for LOC56985 |
|---|
From the 1000 bootstrap trials performed, most branches fall well above a 60% mark, with only one branch within the alphaproteobacteria group falling below the 50% mark. It was very interesting to see that although the Trypanosoma is grouped together with the bacteria it still has a bootstrap value of about 75%.
With the exception of the Trypanosoma, one would say that this protein is only inherited through means of vertical transmission, though given the position and bootstrap value of the Trypanosoma on the protein tree, one may be led to believe that the Trypanosoma has acquired this particular protein through a mode of lateral transmission from the bacteria. Upon further inspection of the protein sequence of the Trypanosoma, one can see that its protein sequence is a lot longer than all the others. A possible explanation for this could be that the gene for this protein has been inserted in the middle of another gene in the Trypanosoma.