LOC56985 Evolution Main: Difference between revisions
No edit summary |
No edit summary |
||
| Line 59: | Line 59: | ||
|} | |} | ||
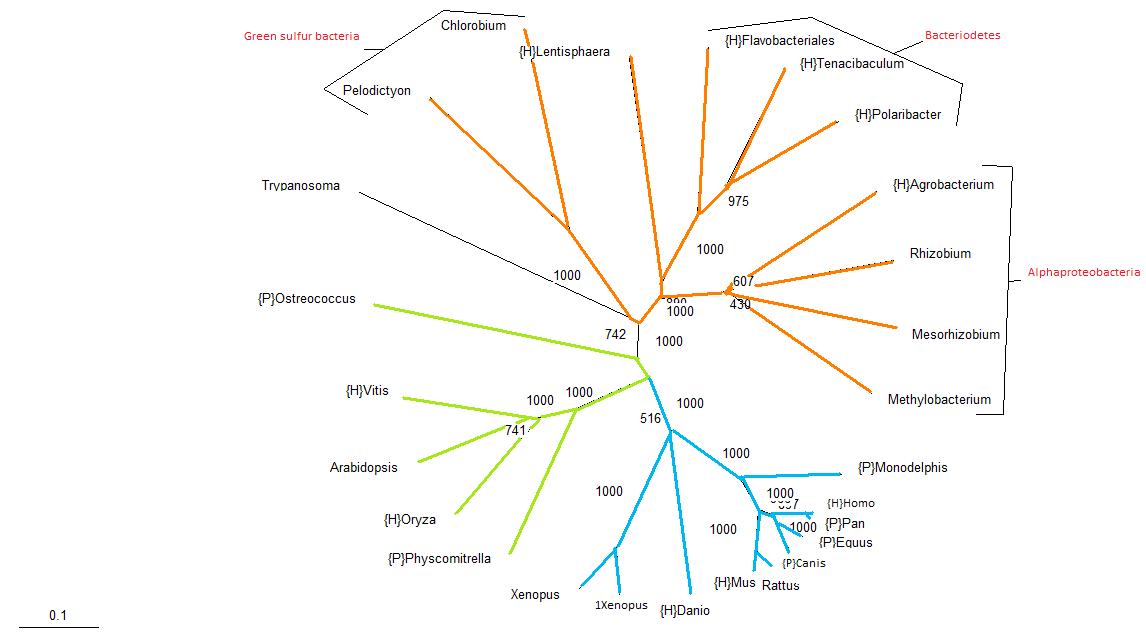
In this tree, bacterial species are shown by orange branches, plants are shown by green branches and vertebrates are shown by blue branches. | In this tree, bacterial species are shown by orange branches, plants are shown by green branches and vertebrates are shown by blue branches. One can see from this tree that the assortment of organisms is generally congruent with traditional phylogenetic trees, with the exception of the ''Trypanosoma'', which is an eukaryote that has been grouped together with the prokaryotes. | ||
One can see from this tree that the assortment of organisms | |||
---- | ---- | ||
[[Hypothetical Protein LOC56985| Back To Main hypothetical protein LOC56985 page]] | [[Hypothetical Protein LOC56985| Back To Main hypothetical protein LOC56985 page]] | ||
Revision as of 22:53, 3 June 2008
BLASTP Search Results
The initial top 60 blast search results, given in FASTA format, can be found in the link below.
For the purpose of creating a phylogenetic tree, sequences from the same species were removed and any sequences containing large gaps in the intial CLUSTAL alignment were also removed. The edited version of the blast search results used for the final clustal alignment can be seen below. Please note that {H} stands for hypothetical protein and {P} stands for predicted protein.
CLUSTALX Alignment
The results for the CLUSTALX alignment can be viewed here.
"The line above the ruler is used to mark strongly conserved positions. Three characters ('*', ':' and '.') are used:
'*' indicates positions which have a single, fully conserved residue
':' indicates that one of the following 'strong' groups is fully conserved:- STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, FYW
'.' indicates that one of the following 'weaker' groups is fully conserved:- CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, FVLIM, HFY
These are all the positively scoring groups that occur in the Gonnet Pam250 matrix. The strong and weak groups are defined as strong score 0.5 and weak score =<0.5 respectively." (Thompson, J.D. et. al. 1997)
Notice the cluster of residues that are invariable (ie '*') across all species. It is likely that these particular residues are important for the Structure of this protein.
Protein Tree
Multiple alignment results from ClustalX were used to create and bootstrap a tree. 1000 bootstrap trials were performed and the results were viewed on Tree View.
| Protein Tree for LOC56985 |
|---|
In this tree, bacterial species are shown by orange branches, plants are shown by green branches and vertebrates are shown by blue branches. One can see from this tree that the assortment of organisms is generally congruent with traditional phylogenetic trees, with the exception of the Trypanosoma, which is an eukaryote that has been grouped together with the prokaryotes.