LOC56985 Function Main: Difference between revisions
From MDWiki
Jump to navigationJump to search
mNo edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image:pfam-a.jpg]] | [[Image:pfam-a.jpg]] | ||
| Line 13: | Line 8: | ||
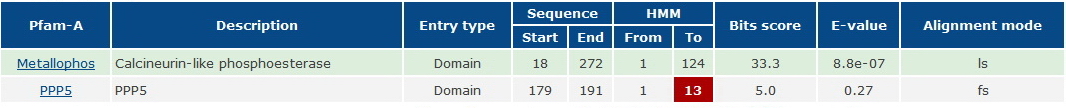
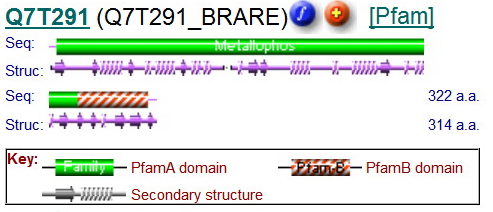
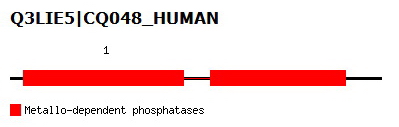
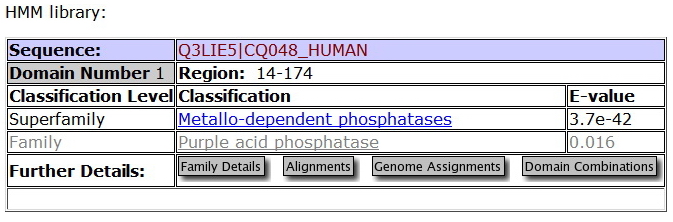
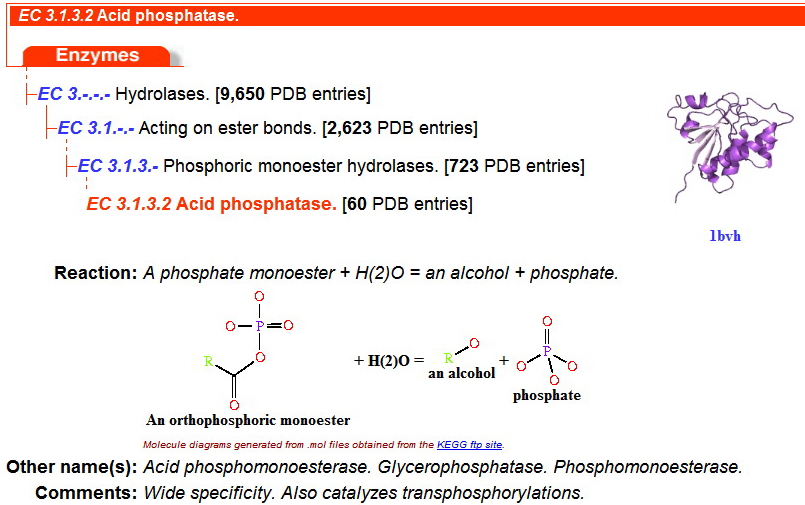
This family was generated automatically from an alignment taken from Prodom 2005.1 (PD344129) subtracting sequence segments already covered by Pfam-A. Sequences from the alignment are LOC56985 orthlogs from Danio rerio, Xenopus laevis, Mus musculus, Oryza Sativa japonica, and Arabidopsis thaliana. | This family was generated automatically from an alignment taken from Prodom 2005.1 (PD344129) subtracting sequence segments already covered by Pfam-A. Sequences from the alignment are LOC56985 orthlogs from Danio rerio, Xenopus laevis, Mus musculus, Oryza Sativa japonica, and Arabidopsis thaliana. | ||
[[Image:metallo.jpg]] | |||
[[Image:super.jpg]] | |||
Revision as of 00:35, 10 June 2008
PfamB PB033851
This family was generated automatically from an alignment taken from Prodom 2005.1 (PD344129) subtracting sequence segments already covered by Pfam-A. Sequences from the alignment are LOC56985 orthlogs from Danio rerio, Xenopus laevis, Mus musculus, Oryza Sativa japonica, and Arabidopsis thaliana.