LOC56985 discussion
Our results provide a structural, phylogenetic, and biochemical basis for the functional annotation of the human gene C17orf48. These findings can also be extrapolated to the multitude of hypothetical metallophosphoesterase orthologs recently identified in vertebrate and plant genomes (Figure 2). High-throughput gene expression data demonstrate highly controlled and tissue specific expression of LOC56985 orthologs, suggesting potential significance to disease states. However, there is minimal experimental evidence concerning LOC56985 orthologs in the literature to ascertain cellular roles and substrate and metal ion specificities by bioinformatical analysis alone.
The Superfamily database classified LOC56985 as a member of the metallo-dependant phosphatase SCOP superfamily but failed to provide a reliable match at the SCOP family level (E-value >0.01), where purple acid phosphatases were the closest match (E-value = 0.016). Pfam identified a metallophosphoesterase domain and classified LOC56985 as a member of the Calcineurin-like phosphoesterase family (PF00149). In fact, different families of the Metallo-dependant phosphatase superfamily are all grouped into the single Calcineurin-like phosphoesterase family according to Pfam. The metallophosphoesterase domain exhibits hydrolase activity (GO:0016787) which classes LOC56985 as an EC class 3 enzyme. Pfam did not detect any additional PAP-type domains (e.g. purple acid phosphatase N-terminal) in LOC56985.
Structural analysis was conducted using the crystal structure of Chain A of an ortholog from Danio rerio, Q7T291 (PDB ID: 2nxf), which was the only solved LOC56985 ortholog structure available at the time of writing. 2nxf shares 54% sequence homology with LOC56985 and coordinates two Zn ions at its active site. Interestingly, the 2nxf sequence contains the Pfam-B domain PB033851 which was automatically generated using an alignment taken from Prodom 2005.1 (PD344129) and subtracting sequence segments already covered by Pfam-A. The PD344129 alignment is entirely made up of sequences orthologous to LOC56985, indicating the possible existence of a novel domain family unique to LOC56985 orthologs.
Comparison of 2nxf structure to all protein structures currently available via the DALI server showed that it is most similar to phosphodiesterases and PAPs. The closest structural homolog was a Glycerophosphodiesterase (2dxl) from Enterobacter aerogenes. The next best matches were iron-binding mammalian PAPs (uteroferrin); 1war from human, 1qhw from rat, and 1ute from pig, in that order. The sixth best match was a cyclic nucleotide phosphodiesterase (2hyp) from Mycobacterium tuberculosis.
All metallo-dependant phosphatases exhibit significant similarities in topology, fold organization active-site organization. They are alpha and beta (a + b) class proteins with a signature βαβαβ secondary structure within a four-layer α/β/β/α sandwich fold. The biochemistry behind metal binding and phosphoesterase activity is similar among all members of the superfamily. Metal ions at the active site are coordinated by conserved loop residues at the carboxy end of parallel beta strands. This structural and functional arrangement has yielded a disperse signature motif in five conserved regions; DX[H/X]-(X)n-GDXX[D/X]-(X)n-GNH[D/E]-(X)n-[G/X]H-(X)n-GHX[H/X]. Each family and subsequently each family member exhibits novel sequence and structural elements based on its metal ion and substrate specificities.
The putative active site of 2nxf is the funnel where PO4 lies bound to a pair of metal ions as shown in the crystal structure. This funnel is the deepest portion of the largest cavity on the protein surface (CastP prediction) and is the putative site of substrate binding by induced-fit. Whilst Zn ions were used as ligands for the dinuclear center during crystallization of 2nxf for structure determination (0.0005 M zinc sulfate was used to supply zinc ions), the metal ions which ligand with 2nxf in vivo was unknown. In fact, the crystal structure shows an additional pair of Zn atoms outside of the dinuclear centre, each bound to a single histidine residue. There is a likelihood that mammalian LOC56985 orthologs are specific for Mn2+, as shown by Canales et al. where the rat ortholog RGD1309906 failed to activate in the absence of Mn2+ (Canales et al. 2008). However, it is not clear whether Mn coordination at the active site or an entirely different interaction (e.g. with the substrate) demonstrated enzyme activation. It is also not clear whether both metal ions at the binuclear center are of the same type or an Fe3+ ion is coordinated alongside a different metal as is the case in some non-mammalian PAPs. If LOC56985 orthologs do indeed exhibit pairs of Zn at the active site as shown for the 2nxf structure, it would be the first instance of a PAP or PAP-like enzyme exhibiting an entirely non-Fe binuclear centre.
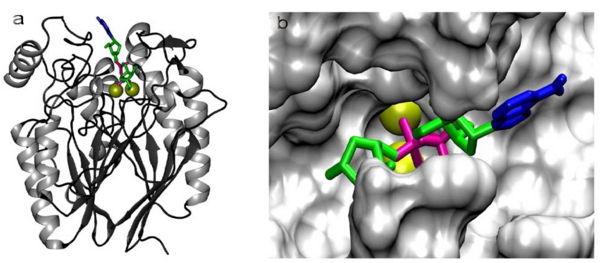
Cavity residues which stabilize the metal ions and bind PO4 are also likely to be involved in the catalytic step. For example, His267 (part of the catalytically relevant GNH[D/E] region in all metallophosphoesterases) interacts with the oxygen of the phosphoanhydride linkage which is duly hydrolysed (Figure 9). Substrate specificity should be accounted for by residues which interact with the non-phosphate regions of the substrate. In a substrate docking model in a homology model generated (using the 2nxf crystal structure) for RGD1309906, a cyclic nucleotide diphosphate sugar substrate (ADP-Ribose) fits into the cavity containing the active center (Figure 9). The beta-phosphate of ADP occupies a position similar to PO4 in the 2nxf structure. The adenine and ribose protrude towards more open areas of the protein surface. It must be noted that the PO4 complexed to 2nxf is not a complete representation of the enzyme-substrate complex but rather a potential representation of PO4 bound to the enzyme subsequent to hydrolysis. PO4 is a considerably smaller molecule than a potential phosphoanhydride substrate. In the true enzyme-substrate complex, specific interactions are likely to depend on an induced fit of the enzyme which PO4 cannot evoke by itself. Substrate binding, catalysis and product release may involve significant conformational changes which cannot be understood from the 2nxf structure.
Multiple sequence alignment of the 2nxf sequence alongside PAP structural homologs (Figure 1) show conservation of the catalytically relevant GNH[D/E] region of metallophosphoesterases but there is relatively minimal conservation across the remainder of the sequence. In contrast, additional conserved patterns are present in all LOC56985 orthologs in both eukaryotes and prokaryotes (Figure 1). Examples of prokaryotic precursors were identified in α-proteobacteria and green sulfur bacteria. Even though these prokaryotic homologs do not exhibit all the conserved patterns seen in eukaryotes, they still exhibit greater sequence homology to LOC56985 than any PAPs. Quantification of residue conservation in the multiple sequence alignment using the Scorecons server returned a result of 99.2%, which rates this alignment as very diverse and informative.
Searching for LOC56985 homologs across complete genomes using TBlastN demonstrated a critical feature of the gene’s evolution. Apart from the above mentioned α-proteobacteria and green sulfur bacteria, orthologs are limited to the protozoan Trypanosoma cruzi, red algae, green algae, mosses, higher plants (tracheophytes), and vertebrates. Orthologs are notably absent in fungi and invertebrates.
On the other hand, human PAP P13686 (1war) is orthologous in a greater variety of organisms; including invertebrates, protozoans, and bacteria, in addition to those in which LOC56985 orthologs are also present. Interestingly, P13686 orthologs are also rare in fungi.
LOC56985 is inherited through vertical transmission, with the only exception seen in T. cruzi. The eukaryotic Trypanosoma was grouped in with the bacteria and still retained a relatively high bootstrap value of 75% in the phylogenetic tree. Further analysis of the Trypanosoma protein sequence showed that it is considerably longer than all others. This finding may lead one to hypothesise about a possible insertion of the protein within another gene in Trypanosoma through means of lateral transmission.
The structural similarities seen between LOC56985 orthologs and PAPs indicate that both protein types likely originated from a gene duplication event in a prokaryotic ancestor. However, the relatively poor sequence similarity observed between LOC56985 orthologs and PAPs, poor HMM matches, phylogenetic differences and possible differences in metal ion specificities indicate that LOC56985 cannot be defined as a typical PAP.
Until very recently, there was no experimental documentation of the cellular role, and substrate and metal ion specificities of LOC56985 orthologs. The only information available at the time of writing was from a recently submitted, unreviewed paper which characterises the product of the orthologous rat gene A9Y0H8 (Canales et al. 2008). This protein has been identified as a Mn2+ dependant ADP-ribose/ CDP-alcohol pyrophosphatase (ADPRibase-Mn). ADP-ribose is a potent intracellular regulator of ion channel activity. Until now, it was assumed that Mg2+ dependant hydrolases in the Nudix superfamily, particularly ADPRibase-I and ADPRibase-II, regulate ADP-Ribose concentration in all mammalian cells. LOC56985 show no sequence or structural similarities to Nudix hydrolases. ADPRibase-Mn was isolated from rat liver supernatants after separation from Nudix hydrolases devoid of CDP-alcohol pyrophosphatase activity. In fact, ADPRibase-Mn is the only known hydrolase which can catalyse hydrolysis of ADP-Ribose as well as CDP-alcohol. CDP-alcohol pyrophosphatase activity may have a role in the biosynthesis pathways of CDP-choline or CDP-ethanolamine. CDP-choline, an intermediate in the generation of phosphatidylcholine from choline, is currently used in the treatment of Alzheimer’s disease as pyschostimulant/ nootropic. Interestingly, the 2nxf crystal structure exhibits a smaller cavity, distinct from the PO4 binding site, where two residues are involved in binding an ethanol molecule. However, sequence alignments fail show to show any conservation of these two residues in other organisms.
High-throughput gene expression data showed that LOC56985 and its orthologs are predominantly expressed in immune tissues in mammals. The restriction of LOC56985 orthologs to vertebrates among animals further supports the probability of an immune specific role. ADPRibase-Mn, and hence orthologs in other vertebrates, may have a signalling role in immune cells where ADP-Ribose acts as a second messenger of stress. ADP-Ribose is an activator of transient receptor potential melastatin channel-2 (TRPM2) which participates in Ca2+ mediated cell death. Immune cells are a major site of TRPM2 expression. Intracellular concentrations of ADP-Ribose increases in response to oxidative or nitrosative stress and trigger opening of TRPM2.
Abstract | Introduction | Results | Discussion | Conclusion | Method | References