LOC56985 results: Difference between revisions
| Line 75: | Line 75: | ||
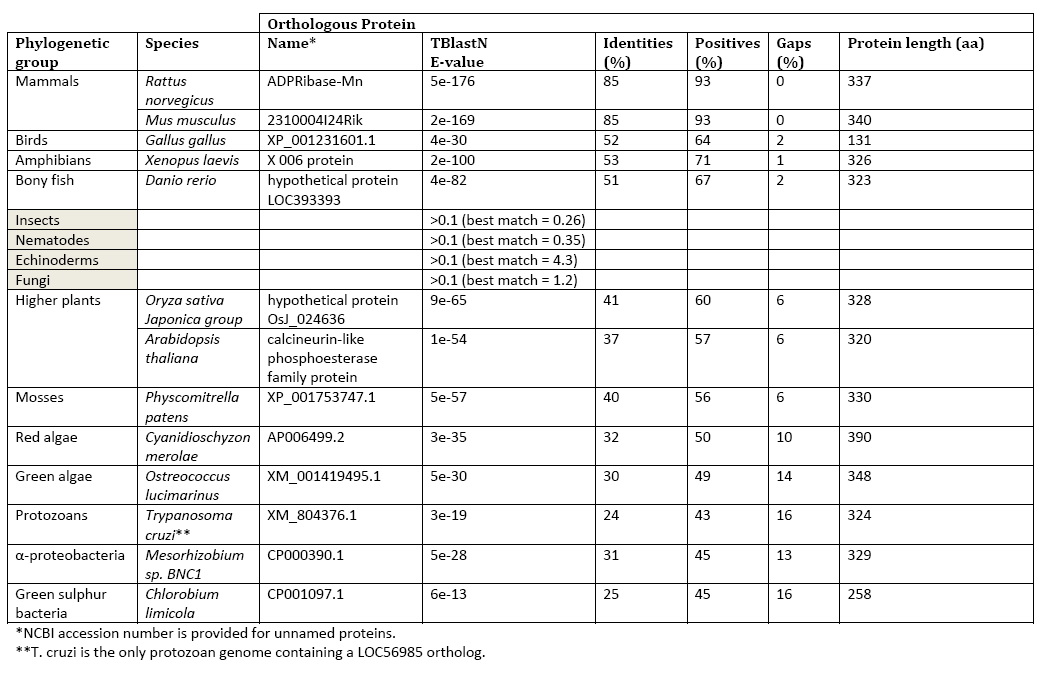
Searching for LOC56985 homologs across complete genomes using TBlastN demonstrated a critical feature of the gene’s evolution. Orthologs are limited to α-proteobacteria, green sulfur bacteria protozoan, protozoan Trypanosoma cruzi, red algae, green algae, mosses, higher plants (tracheophytes), and vertebrates. Orthologs are notably absent in fungi and invertebrates. | Searching for LOC56985 homologs across complete genomes using TBlastN demonstrated a critical feature of the gene’s evolution. Orthologs are limited to α-proteobacteria, green sulfur bacteria protozoan, protozoan Trypanosoma cruzi, red algae, green algae, mosses, higher plants (tracheophytes), and vertebrates. Orthologs are notably absent in fungi and invertebrates. | ||
Table 1: LOC56985 Phylogentics | |||
'''Table 1: LOC56985 Phylogentics''' | |||
[[Image:table.jpg]] | [[Image:table.jpg]] | ||
Revision as of 01:59, 10 June 2008
Evolution
The BlastP search results were mainly comprised of hypothetical or predicted proteins. The closest protein sequence results that weren’t classified as “hypothetical” was the Norway Rat liver manganese (II)-dependent ADP-ribose/CDP-alcohol pyrophosphatase (e-173), the calcineurin-like phosphoesterase family protein of Arabidopsis thaliana (e-53), a Twin-arginine translocation pathway signal in Mesorhizobium bacteria (e-26) and metallophosphoesterases in Rhizobium leguminosarum (e-23), Methylobacterium species (e-21), Pelodictyon phaeoclathratiforme (e -14) and Chlorobium limicola (e-13).
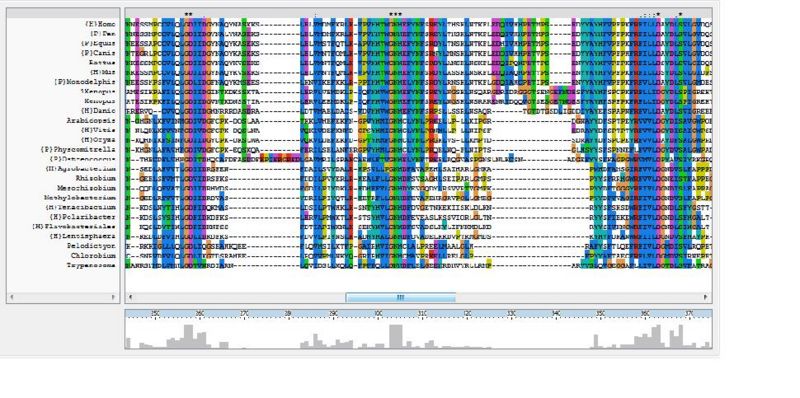
The ClustalX multiple sequence alignment showed various residues that were conserved throughout all species, these are indicated by a '*' above the sequences. These residues are likely play an important role in the structure of the protein.
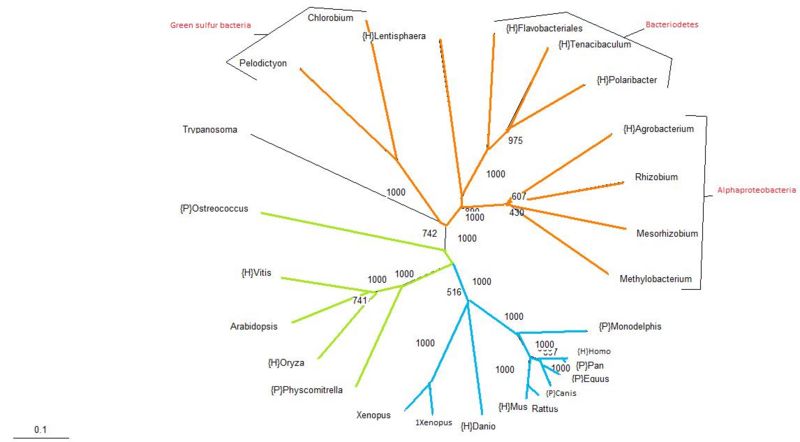
Protein Tree
The protein tree produced from the aligned sequences is analogous with traditional phylogenetic trees, with the exception of the Trypanosoma. Trypanosoma is a parasitic protozoa, which means that it is an eukaryote. In this tree it has been classified along with the bacteria. The bootstrap value for its position in the tree is relatively high at almost 75%, suggesting that perhaps Trypanosoma has acquired this protein by means of lateral transmission. Apart from that single exemplified organism, given the similarity of this protein tree with traditional taxonomy groupings, it seems likely that the usual mode of inheritance for this protein is through vertical transmission.
Bootstrapping values are considerably high, with most branches having a value of greater than 60% and only one branch in the alphaproteobacteria group having less than a 50% confidence.
Structure
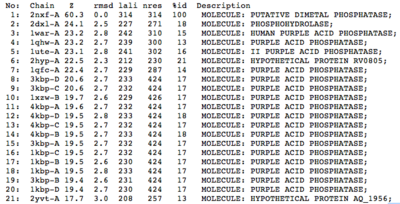
A structural comparison search of the Danio rerio ortholog (2nxf) showed that the gene product had its greatest similarity to a phosphorhydrolase and to various purple acid phosphatases (figure 3). All of these protein matches showed little sequence similarity to the 2nxf however the Z-score of these proteins were significant based on the guide given on the Dali server that Z-scores below 2.5 are insignificant matches. The top hit, 2dxl-A, showed a Z-score of 24.1; other members in the top five matches again showed Z-scores of above 20. Dali works by fitting the two structures together and determining how far apart each of the atoms are from one another to give a root mean square deviation (rmsd), even in the lowest of the top 20 matches the rmsd is 3.0 or below.
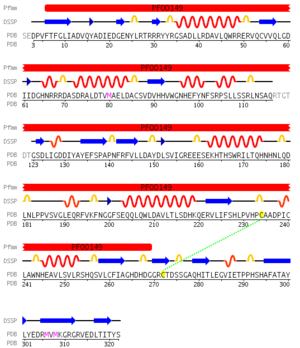
Pfam analysis shows a common domain to all these proteins as being a calcineurin-like phosphoesterase (PF00149). SCOP classification (Murzin et al 1995) of the protein shows it to be an Alpha and Beta (a + b) protein with a 4-layer sandwich fold of alpha/beta/beta/alpha, in the family of puple acid phosphatases (56301). The secondary structure assignment (Kabsch & Sander, 1983) show that the protein contains 20 beta-sheets, 12 alpha-helices and a single disulfide bond between Cys272 and Cys234 (Figure 4); the Pfam domain is shown to stretch from Phe6 to Gly269.
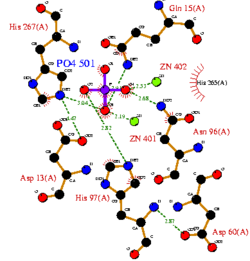
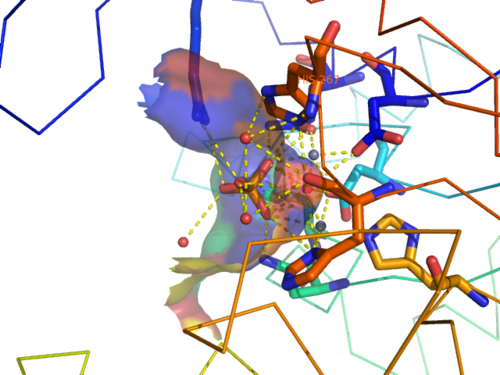
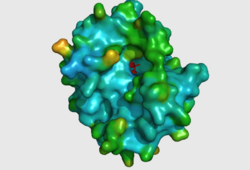
CASTp predicts that the protein contains a number of cavities the largest of which has a volume of 2043.1cubic angstrom covering an area of 950.8squared angstrom. This cavity is very large compared to the others predicted by CASTp. PyMOL analysis of this pocket shows that it is the binding site for the PO4 ligand and contains two Zn atoms buried in the pocket (Figure 5). This site shows three identical residues across the top five protein matches by Dali. This includes the human homolog as well as the Enterbacter aerogenes protein. The sequence of residues is GNH at positions 95 to 97, is unchanged, with a highly conserved acidic amino acid at position 98 down stream of this pattern. The ribbon view (Figure 6) shows that the ligands are positioned at the mid-point sandwiched between the two halves of the protein, above the inner beta-sheet centre of the protein. What only becomes apparent by looking at the surface view of the protein is that this cavity is bridged across the top to form a circle or donut shaped cavity (Figure 5), this may have implications for the type of substrate that can bind to this cavity. A close inspection of the binding site reveals that the GNH pattern appears to be directly involved with ligand binding (Figure 7). The binding site and interactions for the phosphate ion consists of Asp13, Gyn15, Asp60, Asp96, His97, His267 as well as interactions with the two Zn ions. Two residues, Asp96 and His97 form part of the GNH pattern seen in this class of proteins, Gly95 or the acidic amino acid at position 97 does not appear to interact directly with either the Zinc cofactors or the PO4 ligand, it may be that they are important in positioning the other two residues for their bonding interactions or creating the correct fold for the protein.
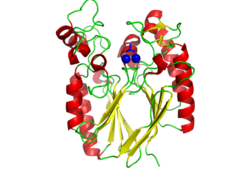
Function
The Superfamily database classified LOC56985 as a member of the metallo-dependant phosphatase SCOP superfamily but failed to provide a reliable match at the SCOP family level (E-value >0.01), where purple acid phosphatases were the closest match (E-value = 0.016). Pfam identified a metallophosphoesterase domain and classified LOC56985 as a member of the Calcineurin-like phosphoesterase family (PF00149). In fact, different families of the Metallo-dependant phosphatase superfamily are all grouped into the single Calcineurin-like phosphoesterase family according to Pfam. The metallophosphoesterase domain exhibits hydrolase activity (GO:0016787) which classes LOC56985 as an EC class 3 enzyme. Pfam did not detect any additional PAP-type domains (e.g. purple acid phosphatase N-terminal) in LOC56985.
The putative active site of 2nxf is the funnel where PO4 lies bound to a pair of metal ions as shown in the crystal structure. This funnel is the deepest portion of the largest cavity on the protein surface (CastP prediction) and is the putative site of substrate binding by induced-fit. Metal ions at the active site are coordinated by conserved loop residues at the carboxy end of parallel beta strands. This structural and functional arrangement has yielded a disperse signature motif in five conserved regions; DX[H/X]-(X)n-GDXX[D/X]-(X)n-GNH[D/E]-(X)n-[G/X]H-(X)n-GHX[H/X]. Each family and subsequently each family member exhibits novel sequence and structural elements based on its metal ion and substrate specificities.
Cavity residues which stabilize the metal ions and bind PO4 are also likely to be involved in the catalytic step. For example, His267 (part of the catalytically relevant GNH[D/E] region in all metallophosphoesterases) interacts with the oxygen of the phosphoanhydride linkage which is duly hydrolysed (Figure 8). Substrate specificity should be accounted for by residues which interact with the non-phosphate regions of the substrate.
Searching for LOC56985 homologs across complete genomes using TBlastN demonstrated a critical feature of the gene’s evolution. Orthologs are limited to α-proteobacteria, green sulfur bacteria protozoan, protozoan Trypanosoma cruzi, red algae, green algae, mosses, higher plants (tracheophytes), and vertebrates. Orthologs are notably absent in fungi and invertebrates.
Table 1: LOC56985 Phylogentics
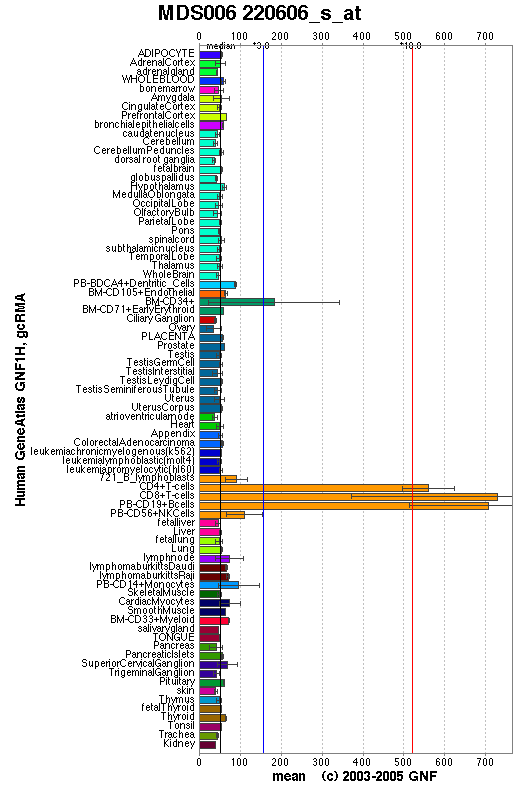
The GeneAtlas dataset for LOC56985 at the GNF database show significantly pronounced expression of LOC56985 in immune cells. The concentration of LOC56985 transcripts in bone marrow CD34+ cells is three times the median value calculated across all tissues. The concentration rises to ten times the median value in CD4+ T cells, CD8+ T cells and peripheral blood CD19+ B cells.
A microarray experiment (listed on the Gene Expression Omnibus (GEO) database, GEO dataset GDS2068), aimed at identifying mouse immune genes from 8734 genomic features showed that the orthologous 2310004I24Rik protein is among 360 which show preferential expression in the thymus, spleen, peripheral blood mononuclear cells, lymph nodes and in vitro activated T cells when compared to non-immune tissues. Unigene’s EST profile viewer showed that LOC66358 is predominantly expressed in the thymus. However, expression levels in spleen samples were not as prominent as was observed in the GEO dataset. Additional evidence which suggest an immune specific role came from fractionation of tissue supernatants from rats to quantitate and compare expression and activity of Nudix hydrolases (ADPRibase-I & ADPRibase-II) and ADPRibase-Mn using northern and acitivity assays (Canales et al. 2008). ADPRibase-Mn expression was significantly higher in thymus and spleen than in non-immune tissues. ADPRibase acitivity was found to be 2.5-5 fold higher in thymus and spleen than liver and muscle, and 4-8 fold higher in splenocytes than in non-immune tissues. Nudix hydrolases did not exhibit an immune specific expression profile.
Abstract | Introduction | Results | Discussion | Conclusion | Method | References