LOC56985 results
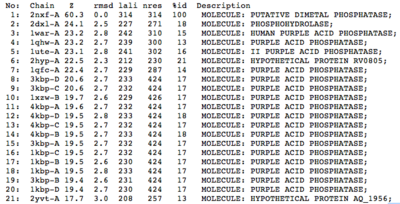
A structural comparison search of the Danio rerio ortholog (2nxf) showed that the gene product had its greatest similarity to a phosphorhydrolase and to various purple acid phosphatases (figure 1). All of these protein matches showed little sequence similarity to the 2nxf however the Z-score of these proteins were significant based on the guide given on the Dali server that Z-scores below 2.5 are insignificant matches. The top hit, 2dxl-A, showed a Z-score of 24.1; other members in the top five matches again showed Z-scores of above 20. Dali works by fitting the two structures together and determining how far apart each of the atoms are from one another to give a root mean square deviation (rmsd), even in the lowest of the top 20 matches the rmsd is 3.0 or below.
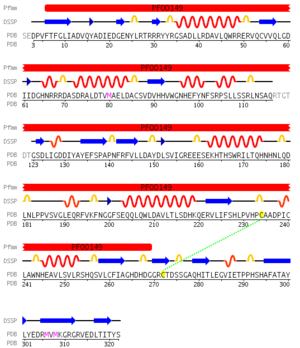
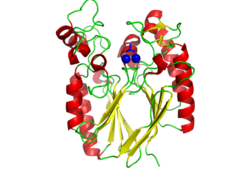
Pfam analysis shows a common domain to all these proteins as being a calcineurin-like phosphoesterase (PF00149). SCOP classification (Murzin et al 1995) of the protein shows it to be an Alpha and Beta (a + b) protein with a 4-layer sandwich fold of alpha/beta/beta/alpha (Figure 4), in the family of puple acid phosphatases (56301). The secondary structure assignment (Kabsch & Sander, 1983) show that the protein contains 20 beta-sheets, 12 alpha-helices and a single disulfide bond between Cys272 and Cys234 (Figure 2); the Pfam domain is shown to tretch from Phe6 to Gly269.
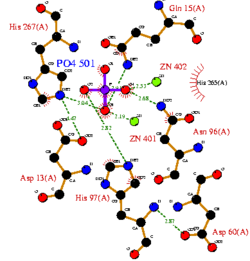
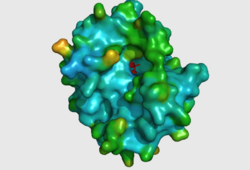
CASTp predicts that the protein contains a number of cavities the largest of which has a volume of 2043.1 covering an area of 950.8. This cavity is very large compared to the others predicted by CASTp. PyMOL analysis of this pocket shows that it is the binding site for the PO4 ligand and contains two Zn atoms buried in the pocket (Figure 3). This site shows three identical residues across the top five protein matches by Dali. This includes the human homolog as well as the Enterbacter aerogenes protein. The sequence of residues is GNH at positions 95 to 97, is unchanged, with a highly conserved acidic amino acid at position 98 down stream of this pattern. The ribbon view (Figure 4) shows that the ligands are positioned at the mid-point sandwiched between the two halves of the protein, above the inner beta-sheet centre of the protein. What only becomes apparent by looking at the surface view of the protein is that this cavity is bridged across the top to form a circle or donut shaped cavity (Figure 3), this may have implications for the type of substrate that can bind to this cavity. A close inspection of the binding site reveals that the GNH pattern does not appear to be directly involved with ligand binding however the acidic amino acid does directly bind to the ligand. It could be that the GNH is integral to creating the correct fold and therefore positioning the acidic amino acid to interact with the ligand.
Abstract | Introduction | Results | Discussion | Conclusion | Method | References