MZ
1. Blast results confirmed human NUBP2 and A.Fulgidus 2PH1 as nucleotide binding proteins.
Psi-blast repeatedly identified nucleotide binding proteins containing the MRP-like ATPase (p-loop), which confirmed the role of NUBP2 and 2PH1 in cell division and signal transduction.
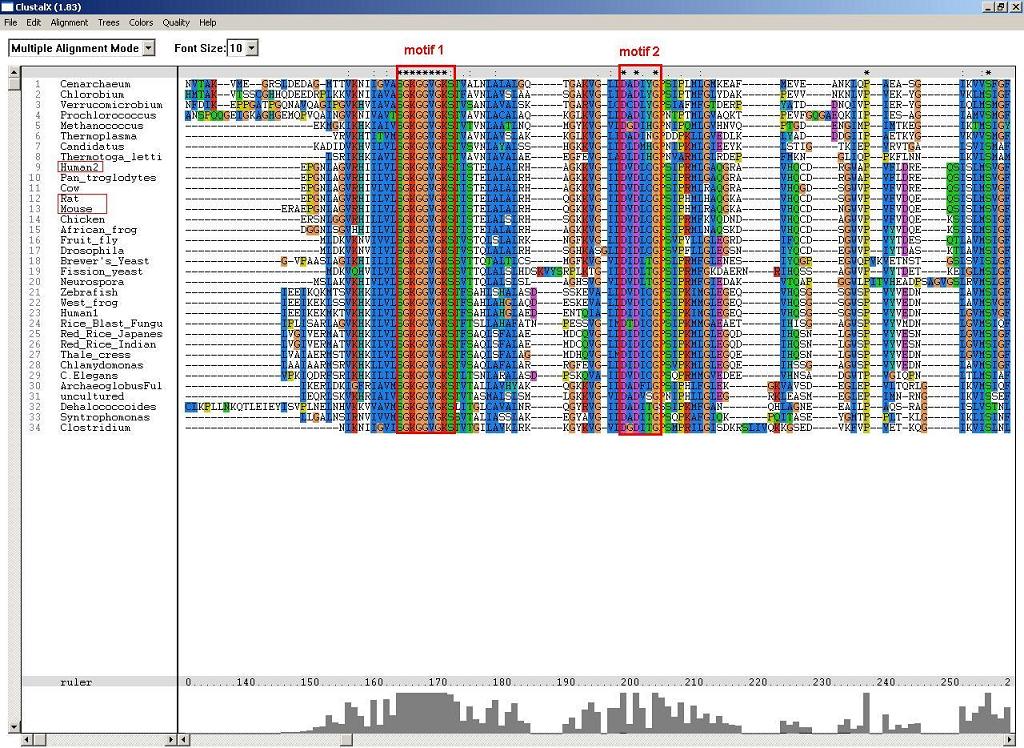
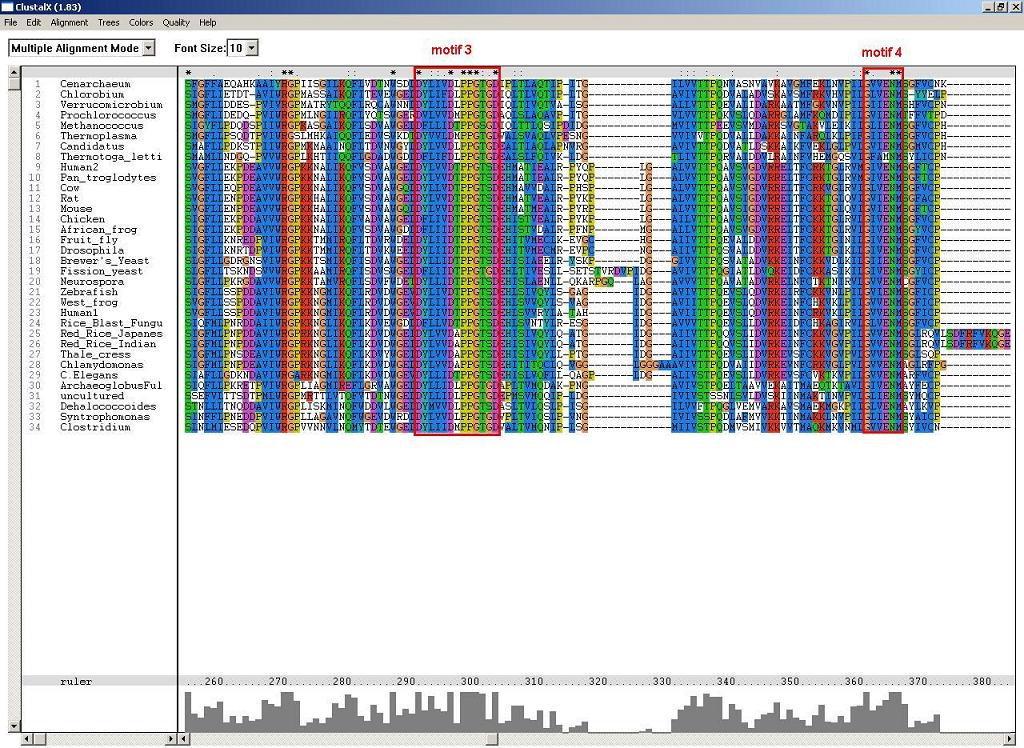
2. ClustalX alignment identified four highly conserved motifs. Firstly, the highly conserved p-loop motif, S-G-K-G(2)-V-G-K-[ST]. Secondly, the overlap motif to the p-loop, D-X-D-[VFLIM]-X-G. Thirdly, the NUBP/MPR motif alpha, D-X-[LIVM]-[LIV]-[LIVF]-D-X-P(2)-G-[TS]-[GS]-D. Fourthly, the NUBP/MRP motif beta, G-[VILF]-[VIA]-X-N-M.
Figure 1. Motif 1 and 2 (p-loop) identified by ClustalX.
Figure 2. Motif 3 and 4 (alpha and beta NUBP/MRP motif) identified by ClustalX.
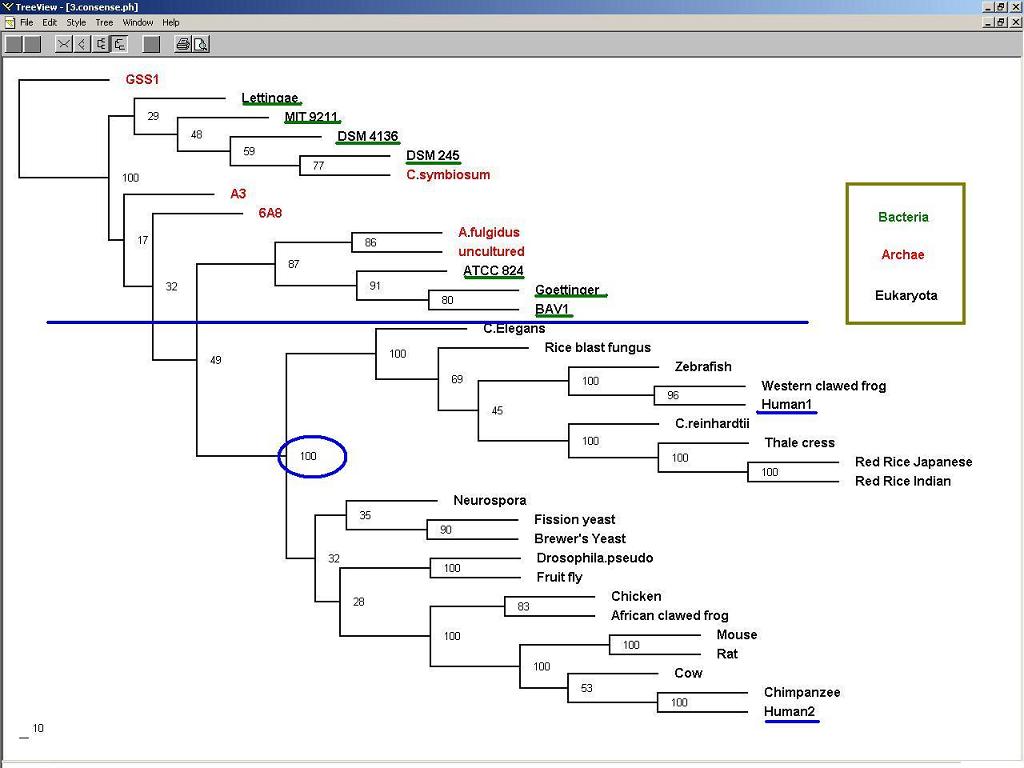
3. The consense Tree
Bacteria and Archae proteins showed weak similarity to Eukaryotes.
Two distinct subgroups were observed in Eukaryota, and each represented by human NUBP1 and human NUBP2:
The NUBP1 group includes C.elegans (flhG protein), rice blast fungus , zebrafish (Nubp1), western clawed frog (LOC100125178), C.reinhardtii, thale chress (NBP), red rice japanse and indian(NBP). In a word, half of the group comes from plants and fungi.
The NUBP2 group includes neurospora (NBP35), yeast (Fe-S clustering binding factor), Drosohpila (CG4858 CG4858-PA), chicken (NUBP2), African clawed frog (MGC78798), mouse (Nubp2), rat (nucleotide-binding protein 2), cow (nucleotide-binding protein 2), chimpanzee (nucleotide-binding protein 2). All the organims in this group belong to yeast and higher eukaryotes, when neurospora has been seen as the link between fungi and yeast.
Figure 3. The consense tree showed the three domains (Red: Archae; Green: Bacteria; Black: Eukaryota) and two distict subgroups (marked by bootstrapped score = 100%) in Eukaryota.