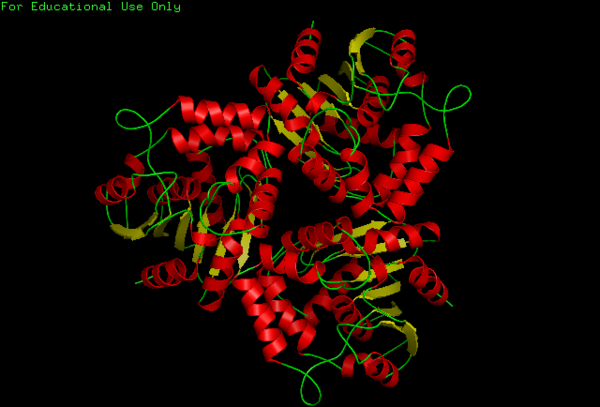
N-myc downstream-regulated gene 2 isoform b
Introduction
The N-myc down regulated gene (NDRG) family belonging to the alpha/beta hydrolase superfamily consists of four members NDRG 1,2 ,3 and 4 where they share 50-60% homology. NDRG is highly conserved in a variety of eukaryotes suggesting functions in cell differentiation and proliferation. Studies have shown that NDRG2 gene is down regulated by Myc proteins via transcriptional repression However, high levels of NDRG2 was observed as Myc expression was reduced in differentiated cells. Myc encodes a transcription factor which binds to DNA of other genes regulating cellular differentiation. Mutations in MYC results in failure of protein to bind correctly to DNA resulting in uncontrolled cell division (cancer).Interestingly, studies have shown that NDRG2 is not repressed by N-myc unlike NDRG1. However, the mechanism in which NDRG2 is not suppressed is still unknown. NDRG2 is a hypoxia responsive gene which induces the apoptosis of tumor cell. To further investigate the structure, function and evolution of NDRG2 an analysis using a range of bioinformatics programs were used.
Methods
Structural analysis of NDRG 2 protein.
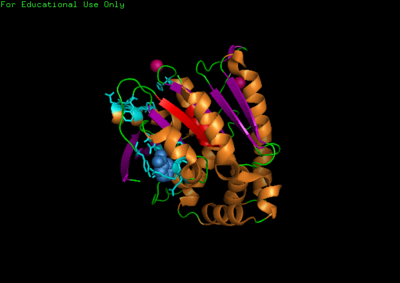
For structure analysis PyMol was used to view PDB structure (2qmq) of N-myc downstream regulated 2. Dali was then used for structural comparison of our protein with other proteins with similar or closely related structure. A secondary structure matching program at EMBL-EBI(http://www.ebi.ac.uk/msd-srv/ssm/cgi-bin/ssmserver) was used to examine our protein from PDB to find similarities within whole SCOP (Structural Classification Of Proteins) archive. Chloroperoxidase protein structure was used as a comparison due to its significant Z-score and low Rmsd value. PyMol was then used to structurally align both proteins together comparing and noting differences in structure.
Determining Function of Protein
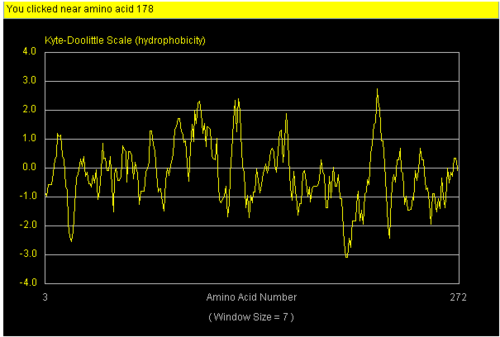
References journals on our protein were found via PUB med. Profunc was use to determine the most likely biochemical function of our protein based on its 3-d structure. Strings database was then used to correlate its interaction with other proteins. Hydropathy plot and hydrophobicity calculator was used to mark specific hydrophobic and hydrophilic region on surface of our protein.
Determining Evolutionary relationship.
PSI-BLAST search was performed using the NCBI web interface (http://www.ncbi.nlm.nih.gov/blast) against the non-redundant database to search for homologous proteins of Human NDRG-2. Amino acid sequences from resulting homologous proteins were saved as FASTA format. The amino acid sequences were then aligned using ClustalX. The alignment was exported as a phylip file extension and subsequently used in creating a phylogenetic tree using Phylip software.
Results
The NDRG2 molecule was related to the a/b hydrolase superfamily with key a characteristic where the second B-sheet of the molecule is anti-parallel compared to the rest of the B-sheet.
This highly conserved region is located at the core of the NDRG2 protein linked to ligands directly to the surface of the protein.
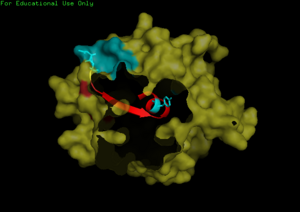
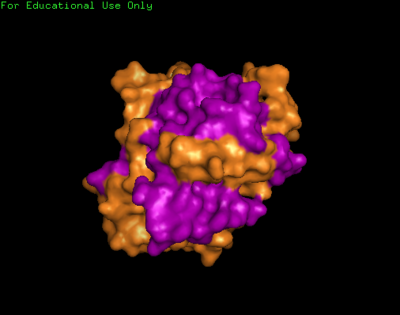
We found a region with hydrophobic characteristic situated at the surface of the protein which plays a role in its translocation from cytoplasm into the nucleus. In which the hydrophobis region binds to nuclear membrane which facilitates its diffusion into the nucleus.
A single Chloroperoxidase domain was found to be most closely related to NDRG2 structure where each of its domains best matches overall structure of our protein based on RMSD (Root Mean Square Deviation) values.
Discussion
Discussion: Function of Chloroperoxidase is to hydrolyze hydrogen peroxide as high concentrations of it have negatives effects in cells. E.g. accumulation of H2O2 In plants cells have damaging effects DNA, lipid membranes and organelles and plants rely on these enzymes to prevent accumulation of H2O2. However, presence of H2O2 also plays a role in plant defenses against pathogens. Based on this, NDRG2 function in the regulation of H2O2 levels within a cell.
Believed to play a role in the regulation of reactive oxygen species. FigureX shows overlay of human NDRG-2 protein with strep L. This is due to low RMSD values indicating the best superimposition between both 3-d structures. Based on homology of both structure, this molecule may have the same or very similar function.
The presences of many hydrophobic residues on the surface of NDRG-2 shows that it is needed to facilitate entry pass the nuclear membrane into the nucleus. Hydrophobic region binds to hydrophobic nuclear membrane regions via hemophilic interaction which then facilitates its endocytosis into the nucleus.Within this region consists of ligands which connects directly to the conserved region core of the protein which may also play a role in faculitating its translocation into the nucleus.
Based on structure of chloroperoxidase NDRG-2 protein could be active as a tetramer this suggests that more than molecule is required to form a full functional molecule where the active site can be formed when multiple domains are brought together.