Phostphatase: Difference between revisions
No edit summary |
No edit summary |
||
| (9 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<font size = "4">'''Phosphatase'''</font> | |||
[[Image:Document9_01.png]] | [[Image:Document9_01.png]] | ||
| Line 13: | Line 15: | ||
MSA corelate with with study done by Maliekal ''et al'' (2006) | |||
[[Image:Document19_08.png]] | |||
Alignment of rat and human Neu5Ac-9-P phosphatase with homologous sequences. The following sequences are aligned: ''Rattus | |||
norvegicus'' (Rnor, gi-34859431), ''Homo sapiens'' (Hsap, gi-23308749), ''Xenopus laevis'' (Xlae, gi-46250196), ''Danio rerio'' (Drer, gi- | |||
63101958), and ''Drosophila melanogaster'' (Dmel, gi-28381565). Only the first 280 residues of the latter sequence are shown. Completely | |||
conserved residues are shown in boldface type. Asterisks indicate the extremely conserved residues in phosphatases of the HAD family <sup>8</sup>. | |||
*N-acetylneuraminic acid phosphatase orthologs shared 3 motifs found in phosphatases | |||
**1<sup>st</sup> motif comprising two extremely conserved aspartates (D) | |||
**2<sup>nd</sup> motif comprising a conserved serine (S) or threonine (T) | |||
**3<sup>rd</sup> motif comprising a conserved lysine (K) and two conserved aspartates (D) | |||
[[Role in Human|Next Page]] | [[Role in Human|Next Page]] | ||
Latest revision as of 01:37, 12 June 2007
Phosphatase
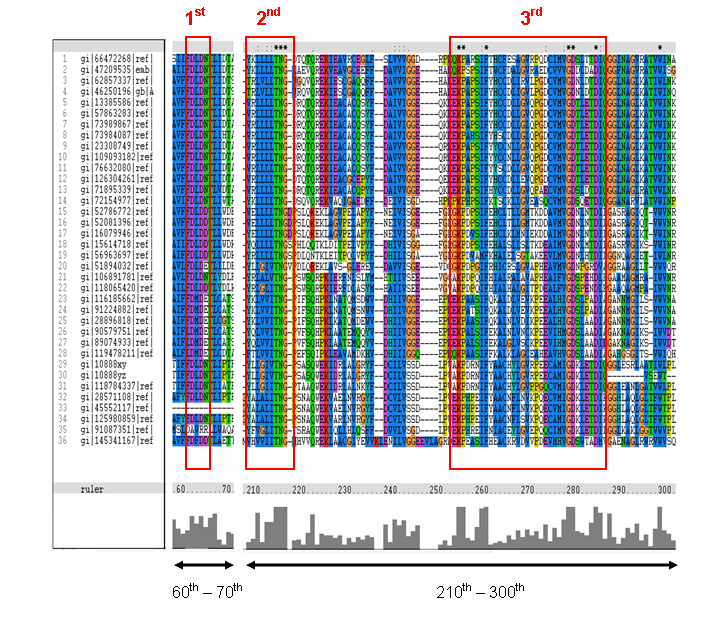
MSA of the 2gfh with 35 others proteins. Only the 60th – 70th and the 210th -300th amino acid
sequence were shown to illustrate the conserved and invariant regions. The 3 boxed-up sequences were either conserved or invariant regions.
- 1st - aspartic acid (D)
- 2nd - threonine (T), asparagine (N) and glycine (G)
- 3rd - lysine (K) and aspartates (D)
MSA corelate with with study done by Maliekal et al (2006)
Alignment of rat and human Neu5Ac-9-P phosphatase with homologous sequences. The following sequences are aligned: Rattus
norvegicus (Rnor, gi-34859431), Homo sapiens (Hsap, gi-23308749), Xenopus laevis (Xlae, gi-46250196), Danio rerio (Drer, gi-
63101958), and Drosophila melanogaster (Dmel, gi-28381565). Only the first 280 residues of the latter sequence are shown. Completely
conserved residues are shown in boldface type. Asterisks indicate the extremely conserved residues in phosphatases of the HAD family 8.
- N-acetylneuraminic acid phosphatase orthologs shared 3 motifs found in phosphatases
- 1st motif comprising two extremely conserved aspartates (D)
- 2nd motif comprising a conserved serine (S) or threonine (T)
- 3rd motif comprising a conserved lysine (K) and two conserved aspartates (D)