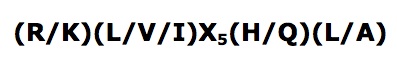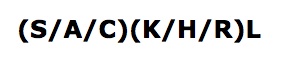Phytanoyl-CoA dioxygenase Function
Interpro Analysis
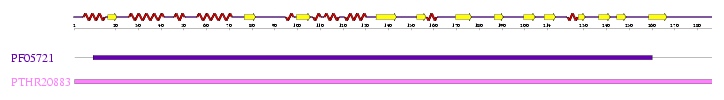
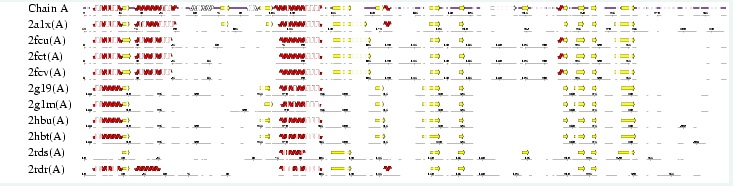
Interpro analysis showed motif similarity to the family of phytanoyl-CoA hydroxylases (PhyH) (Figure 1) and Secondary Structure analysis via profunc showed high structural identity to phytanoyl-CoA hydroxylase (PDB code; 2a1x) (Figure 2). Although the sequence homology between Phytanoyl-CoA dioxygense and the hydroxylase identified was only 24.9%, a pymol analysis showed the two enzymes to have a high structural similarity.
Phytanoyl-CoA hydroxylase
Phytanoyl-CoA hydroxylase is an enzyme catalysing the first step of alpha oxidation in the peroxisome (figure 3). It functions as an oxidoreductase to oxidise phytanoyl-CoA to 2-hydroxyphytanoyl-CoA. 2-oxoglutarate is simultaniously reduced to succinate and carbon dioxide.
The pathway of alpha oxidation in the peroxisome. (Image source http://www.biochemsoctrans.org
Alpha oxidation is a series of reactions in the peroxisome that remove the alpha carbon from uneven, long-chain fatty acids. The products of alpha oxidation are fed into the peroxisomal beta oxidation pathway before transport to the mitochondria for further beta oxidation to acetyl-CoA. Acetyl-CoA is the entry product into the tricarboxylic acid (TCA) cycle of energy production.
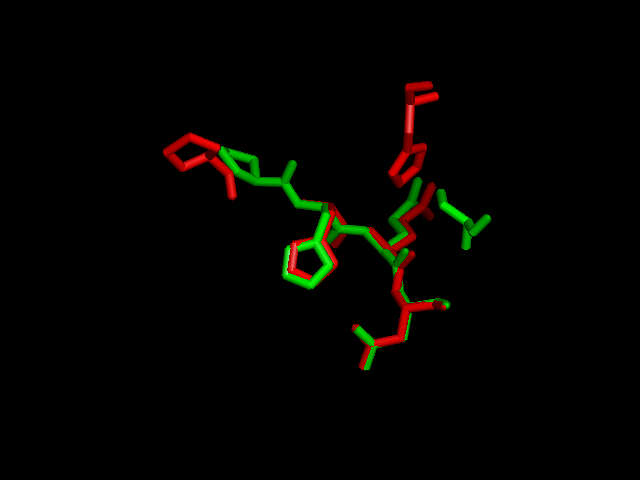
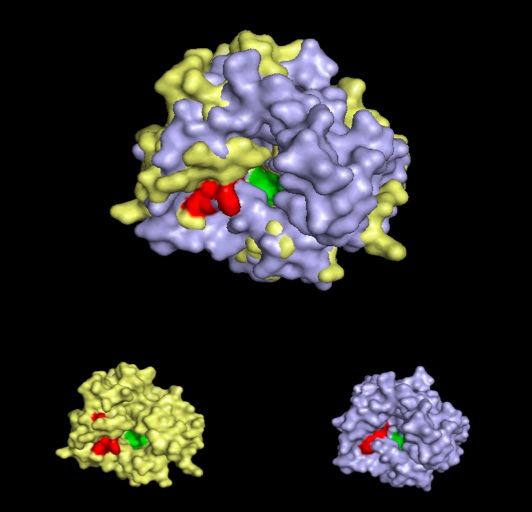
Pymol analysis of similarity between Phytanoyl-CoA dioxygenase and Phytanoyl-CoA hydroxylase
Inferance of protein function is made by comparison of structure between phytanoyl-CoA dioxygenase and phytanoyl-CoA hydroxylase. Although not sequentially similar (Blast score? Clustalx score?) a pymol alignment highlights the structural similarities.
Fe2+ binding sites
A putative iron binding site was identified in phytanoyl-CoA dioxygenase by structural comparison to the iron binding site of phytanoyl-CoA hydroxylase. Residues PRO155, HIS156, GLN157 and ASP158 show structural similarity to the PRO173, HIS175, GLN176 and ASP177 of the hydroxylase enzyme. HIS220 is also involved in iron binding in the hyroxylase but no histadine residue exist in this position in the dioxygenase. It is hypothesised that the histadine binding is replaced by a serine at position 160, which is spacially close to the histadine in the pymol alignment.
2-oxogluatarate binding site
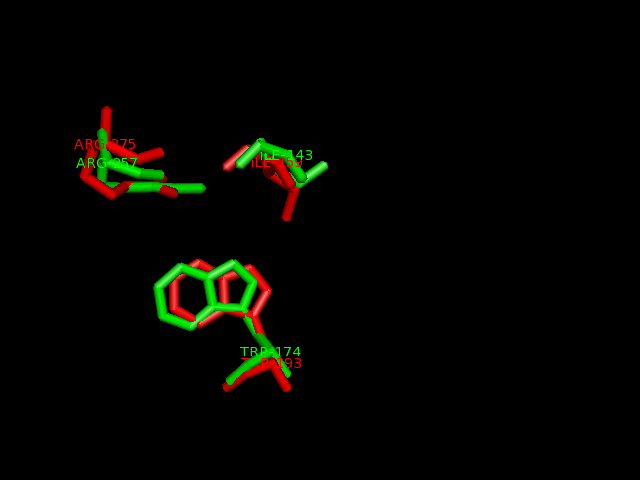
Ligand binding template results from profunc matched TRP193, ILE159 and ARG275 from hydroxylase with the TRP174, ILE143 and ARG257from the dioxygenase (e-value 1.95x10-8).
Pymol alignment of phytanoyl-CoA hydroxylase and phytanoyl-CoA dioxygenase
A pymol alignment of phytanoyl-CoA hydroxylase and phytanoyl-CoA dioxygenase showed conserved structural positions of both the iron binding site and the 2-oxogluterate binding site in the largest cleft of the dioxygenase enzyme. This is the probable active site due to the presence of both putative binding sites and good accessibility for substrates (Cleft analysis results).
Peroxisomal Targeting
A STRING analysis (http://string.embl.de/) failed to find any interacting partners for phytanoyl-CoA dioxygenase and the absense of homology with phytanoyl-CoA hydroxylase prevent the inference of interactions.
Phytanoyl-CoA hydroxylase interacts with the membrane-bound peroxisomal transporter protein PEX-7, which recognises the type 2 peroxisomal targeting signal at the N-terminus. After transport into the peroxisome, the N-terminus is cleaved between Thr30 and Ser31 to produce the mature protein.
The pymol analysis indicates that phytanoyl-CoA dioxygenase may have the same function as phytanoyl-CoA hydoxylase due to the structural similarities and conserved ligand binding sites. Phytanoyl-CoA hydroxylase functions in the peroxisome to catalyse the first step of alpha oxidation and is synthesed as a pro-protein with a N-terminal type-2 peroxisomal targeting sequence (PTS). The targeting sequence is recognised by the peroxisomal receptor protein PEX7 for transport into the peroxisome. Once inside the peroxisome the N-terminus PTS-2 is cleaved between residues THR30 and SER31 to produce the mature phytanoyl-CoA hydroxylase (McDonough, M. 2005). However, phytanoyl-CoA dioxygenase does not contain an N-terminus PTS-2 or a PTS-1 at the carboxyl end. This suggests that the dioxygenase enzyme does not utilise the same receptor for entry into the peroxisome, but may enter by an undetermined mechanism. Dyer et al., 1996 described peroxisomal proteins that do not contain either a PTS-1 or PTS-2 sequence.
Pathology
Mutations in phytanoyl-CoA hydroxylase have been shown to cause Refsum’s disease (RD), charecterised by retinitis pigmentosa, anosmia, sensory neuropathy and phytanic acidaemia in adulthood. The symptoms of RD arise due to the build up of phytanoyl-CoA, which is inhibitory to some mitochondrial biochemical pathways.
Pyruvate dehydrogenase catalyses the production of acetyl-CoA from pyruvate for entry into the TCA cycle. The TCA cycle produces FADH2 and NADH as substrates for the electron transport chain. Acetyl-CoA is produced from pyruvate in a reaction catalysed by the pyruvate dehydrogenase complex.
The pyruvate dehydrogenase complex is composed of the three enzymes (E1, E2 and E3) that produce acetyl-CoA in three defined, sequential reactions. E1 is inhibited by high concentrations of phytanoyl-CoA reducing the input of substrate into the TCA cycle.
The 2-oxoglutarate dehydrogenase complex is similar in structure to the pyruvate dehydrogenase complex and also contains three enzymatic subunits. E1 of the 2-oxoglutarate complex is also inhibited by phytanoyl-CoA. This situation is benificial to a cell with normally functioning phytanoyl-CoA hydroxylase as 2-oxoglutarate is an essential reactant in the oxidation of phytanoyl-CoA.
The result of inhibition of 2-oxoglutarate- and pyruvate dehydrogenase is a reduction in TCA cycle activity and therefore reduced ATP production. The high concentrations of phytanoyl-CoA dioxygenase in neurons may account for the neurotoxicity of RD.
Expression
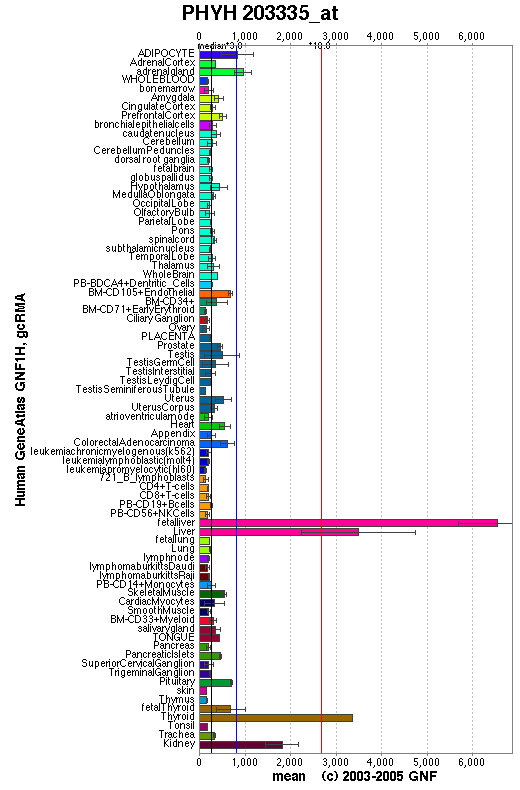
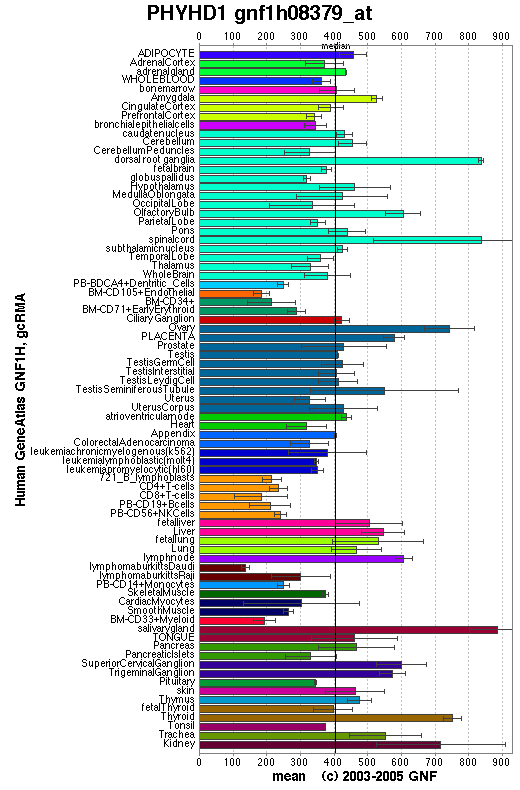
An expression profile created from the human gene atlas (http://symatlas.gnf.org) shows that phytonoyl-CoA dioxygenase (figure 10) is expressed in different tissues to phytanoyl-CoA hydroxylase (figure 11). There is increased expression of the dioxygenase enzyme in the spinal cord, dorsal ganglion and olfactory bulb whereas the hydroxylase enzyme has increase expression in adipocytes and hepatocytes.
Interestingly, it is phytanoyl-CoA dioxygenase that is expressed highly in the tissues that degenerate in Refsum's disease. The dorsal root ganglia and and the spinal cord are involve in the processing of signals from sensory neurons and the olfactory bulb senses odours. These areas are affected by Refsum's disease which is charecterised by sensory neuropathy and anosmia. High expression of the phytanoyl-CoA hydroxylase in hepatocytes and adipocytes might be related to the phytanic acidaemia of Refsum's disease. Phytanic acidaemia may cause the other symptoms.
Increase expression of the dioxygenase enzyme in the dorsal root ganglion, olfactory bulb and spinal cord may indicate a need for increased processing of phytanoyl-CoA. Therefore, it is hypothesised that when the hydroxylase enzyme is non-functional causing phytanic acidaemia, it is these tissues that are most affected.