Phytanoyl-CoA dioxygenase Function
'Pymol analysis of similarity between Phytanoyl-CoA dioxygenase and Phytanoyl-CoA hydroxylase
Inferance of protein function is made by comparison of structure between phytanoyl-CoA dioxygenase and phytanoyl-CoA hydroxylase. Although not sequentially similar (Blast score? Clustalx score?) a pymol alignment highlights the structural similarities.
Fe2+ binding sites
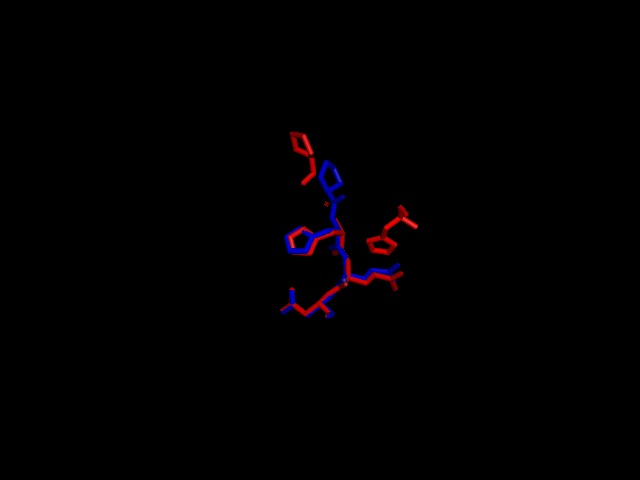
A putative iron binding site was identified in phytanoyl-CoA dioxygenase by structural comparison to the iron binding site of phytanoyl-CoA hydroxylase. Residues PRO155, HIS156, GLN157 and ASP158 show structural similarity to the PRO173, HIS175, GLN176 and ASP177 of the hydroxylase enzyme. HIS220 is also involved in iron binding in the hyroxylase but no histadine residue exist in this position in the dioxygenase. It is hypothesised that the histadine binding is replaced by a serine at position 160, which is spacially close to the histadine in the pymol alignment.
Figure 3; a pymol alignment of the recognised iron binding site of phytanoyl-CoA hydroxylase (Red) with the putative iron binding site of phytanoyl-CoA dioxygenase (Blue)
2-oxogluatarate binding site Ligand binding template results from profunc matched TRP174, ILE143 and ARG257 from hydroxylase with the TRP193, ILE159 and ARG275 from the dioxygenase (e-value 1.95x10-8).
Aligment of Phytanoyl-CoA hydroxylase and Phytanoyl-CoA dioxygenase