Pyridoxal Phosphatase Results: Difference between revisions
No edit summary |
No edit summary |
||
| Line 63: | Line 63: | ||
[[Image:Cleft Analysis PyMOL 2cfsA V2.jpg]][[Image:Cleft Analysis PyMOL 2oycA V2.jpg]] <BR> | [[Image:Cleft Analysis PyMOL 2cfsA V2.jpg]][[Image:Cleft Analysis PyMOL 2oycA V2.jpg]] <BR> | ||
'''Active Site of 2cfsA''' <BR> | |||
[[Image:2CFS_Active_Site.jpg]] | |||
'''PROFUNC''' <BR> | '''PROFUNC''' <BR> | ||
Revision as of 02:08, 31 May 2008
Evolution
Structure
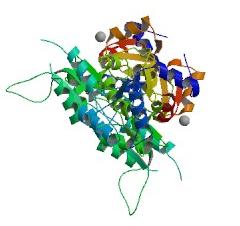
Crystal structure of the target protein (2cfsA), as obtained from the Protein Data Bank (PDB)
Based on the information provided in the website, Pyridoxal Phosphatase has the following characteristics:
- Pyridoxal Phosphatase is isolated from Homo Sapiens and is expressed in Escherichia Coli.
- Structure is similar to the Pyridoxal Phosphate Phospatase protein
- 1 (A) Chain
- Consists of Magnesium components
- Resolution of 2.40 angstroms. This means that the number of sidechains in the wrong rotamer is smaller as compared to proteins of a higher resolution (>2.5 angstroms). Other characteristics of proteins of similar resolutions are: (1) many small detectable errors, (2) correct folding, (3) fewer number of errors in the surface loops and (4) visible water molecules and small ligands.
http://www.rcsb.org/pdb/explore/explore.do?structureId=2cfs
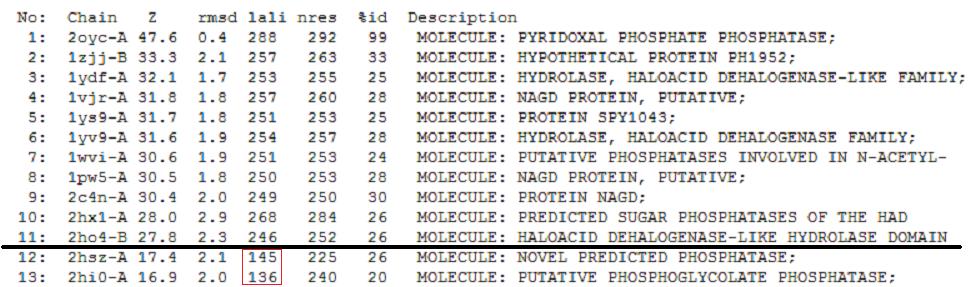
DALI
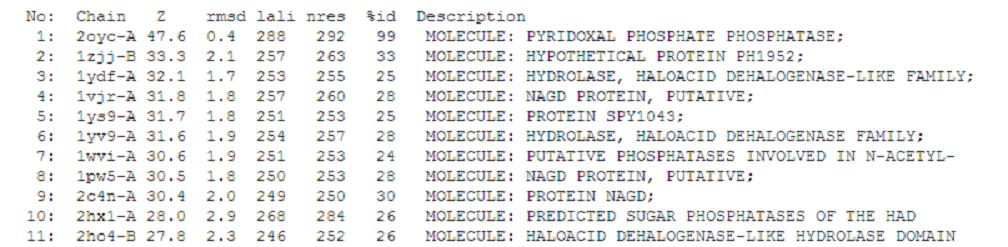
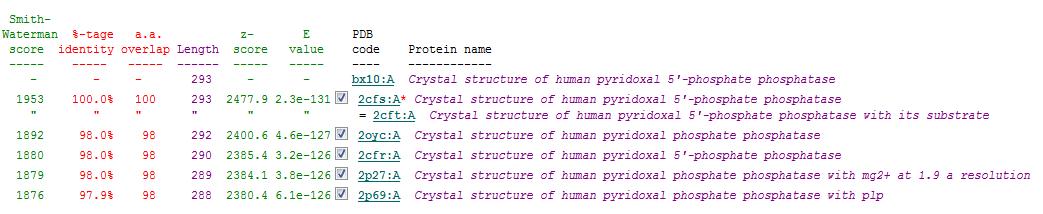
A total of 176 hits were generated, of which only the first 11 (as shown above) were predicted to be significant. The others were rejected on account that their respective nres scores (refer to the red, boxed section in the diagram below) were less than half of Pyridoxal Phosphatase's (nres: 296).
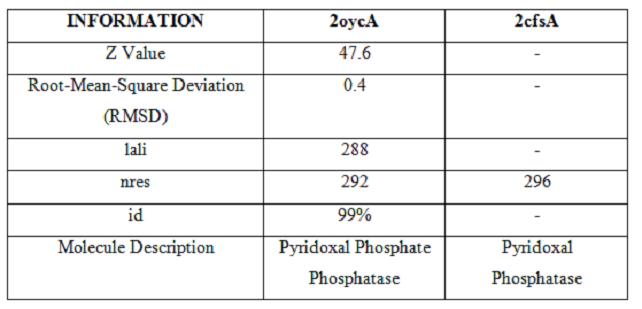
It was noted that none of the hits actually matched Pyridoxal Phosphatase. The closest hit was a Pyridoxal Phosphate Phosphatase (PDB ID 2oycA) which bore great similarity to the protein of interest. Below is a table comparing both Pyridoxal Phosphatase (2cfsA) and the Pyridoxal Phosphate Phosphatase (2oycA)
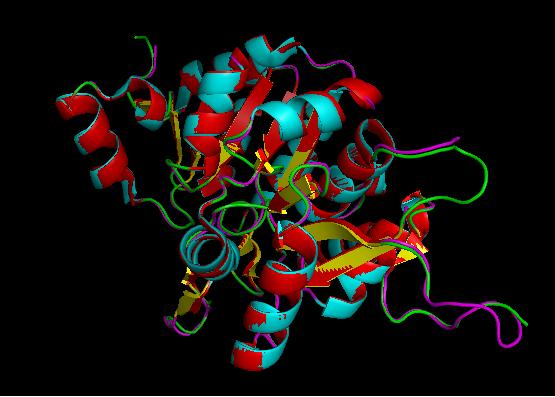
Using the PyMOL software, 2oycA was superimposed against 2cfsA, and both structures, as shown below, are structurally similar.
PDBsum
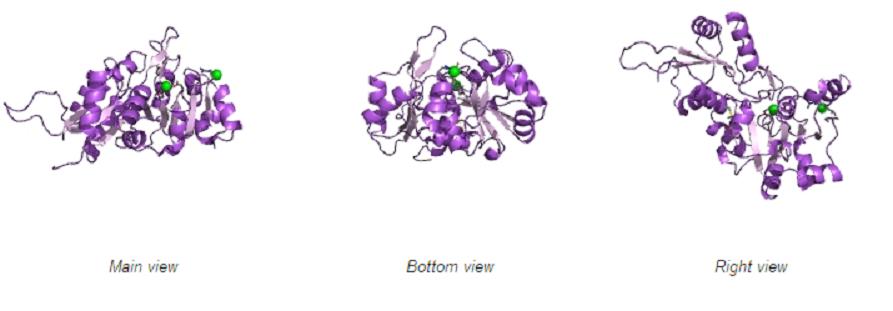
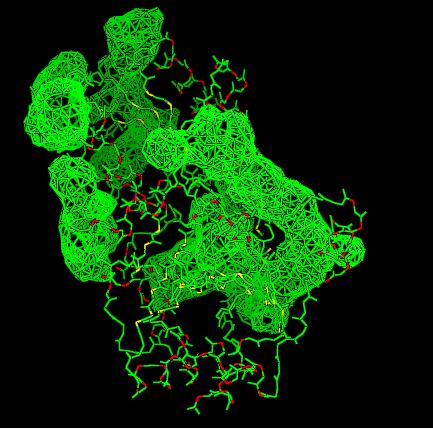
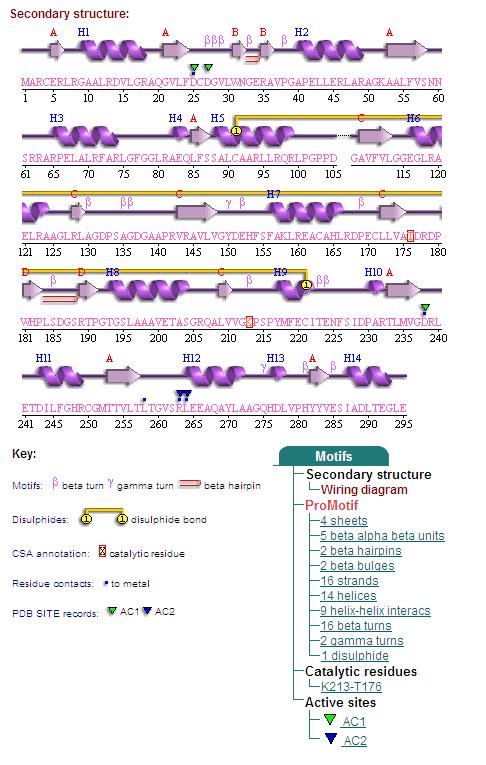
Different views of 2cfsA. The purple chains represent the amino acid chain of 293 amino acids while the green spheres represent the magnesium ions (x2).
By clicking the "Protein chain" link, the user was re-directed to a website containing information pertaining to the secondary structures of both 2cfsA and 2oycA.
Secondary Structures of 2cfsA (L, http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl) and 2oycA (R)
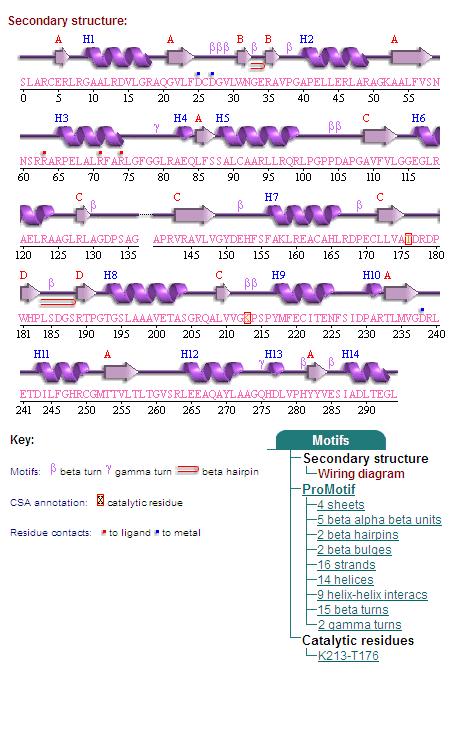
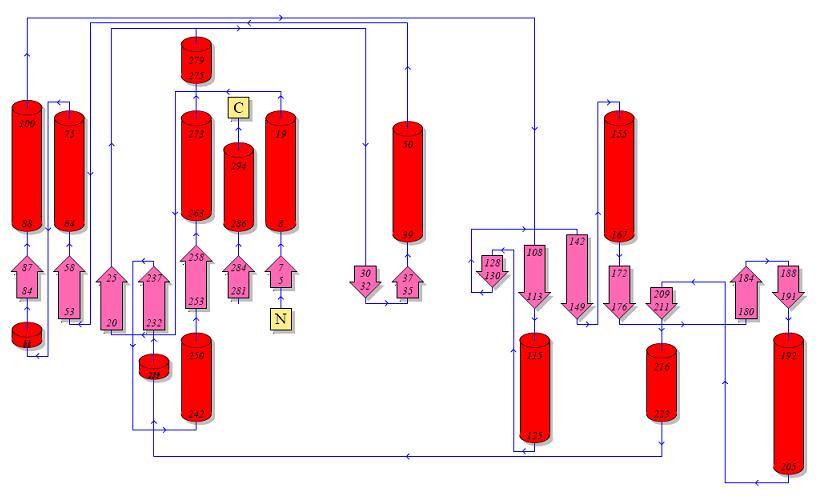
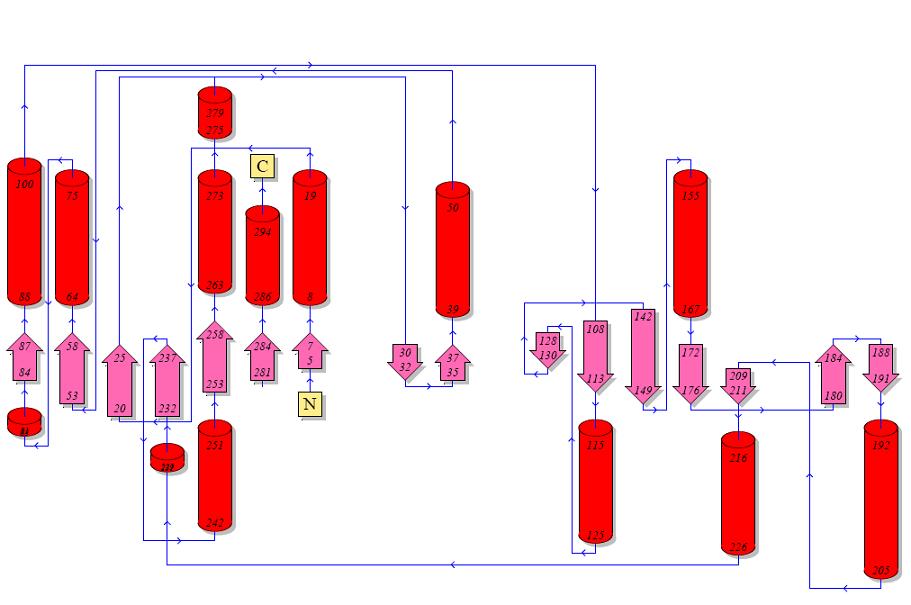
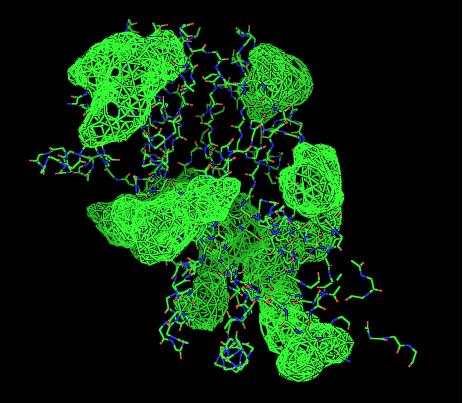
Topology diagrams of 2cfsA (Top, http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=2cfs&template=protein.html&r=wiring&l=1&chain=A) and 2oycA (Bottom).
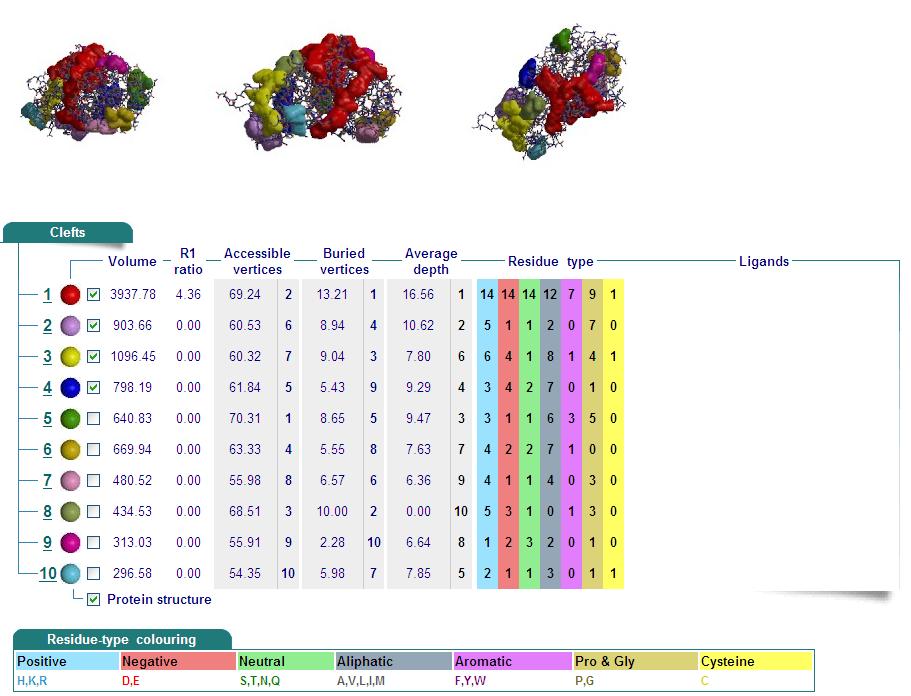
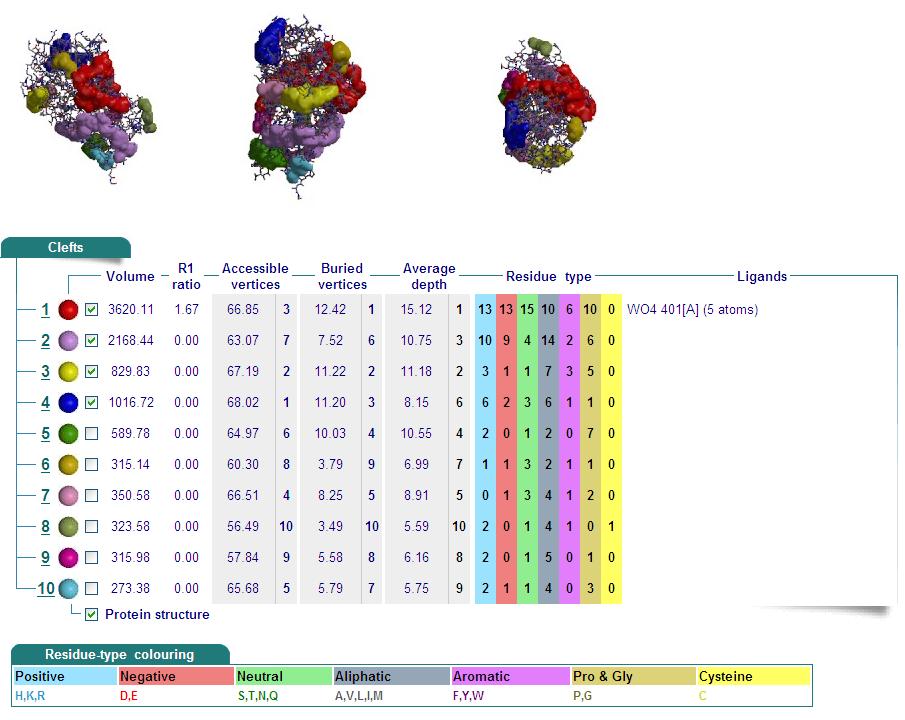
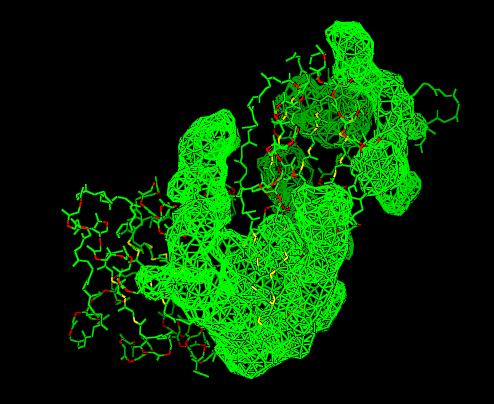
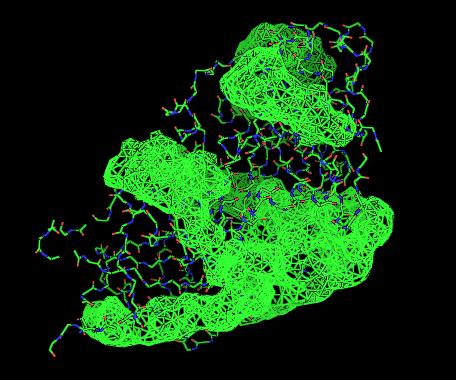
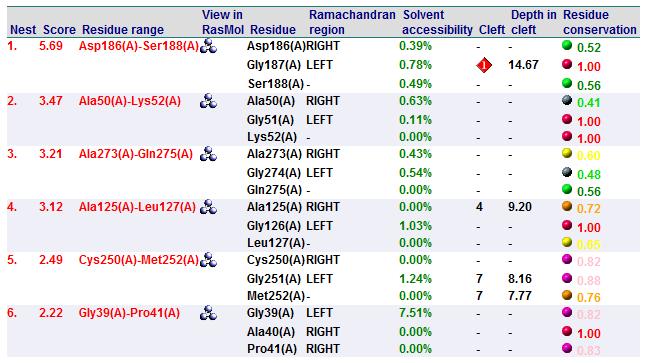
Cleft Analyses of 2cfsA (Top, http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=2cfs&template=clefts.html&pdbcode=2cfs&r=speedfill) and 2oycA (Bottom).
Cleft Analysis via PyMol
- VIEW 1: 2cfsA (L) and 2oycA (R)
- VIEW 2: 2cfsA (L) and 2oycA (R)
Active Site of 2cfsA
File:2CFS Active Site.jpg
PROFUNC
Related Protein Sequences in the PDB (SAS)

Matches to existing PDB Structures
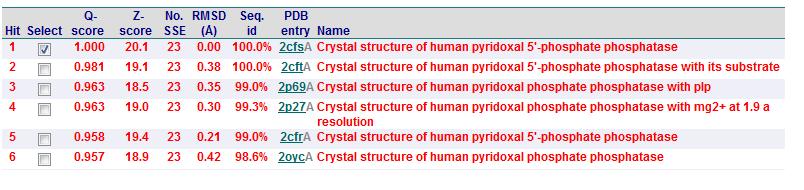
Secondary Structure Matching (SSM)
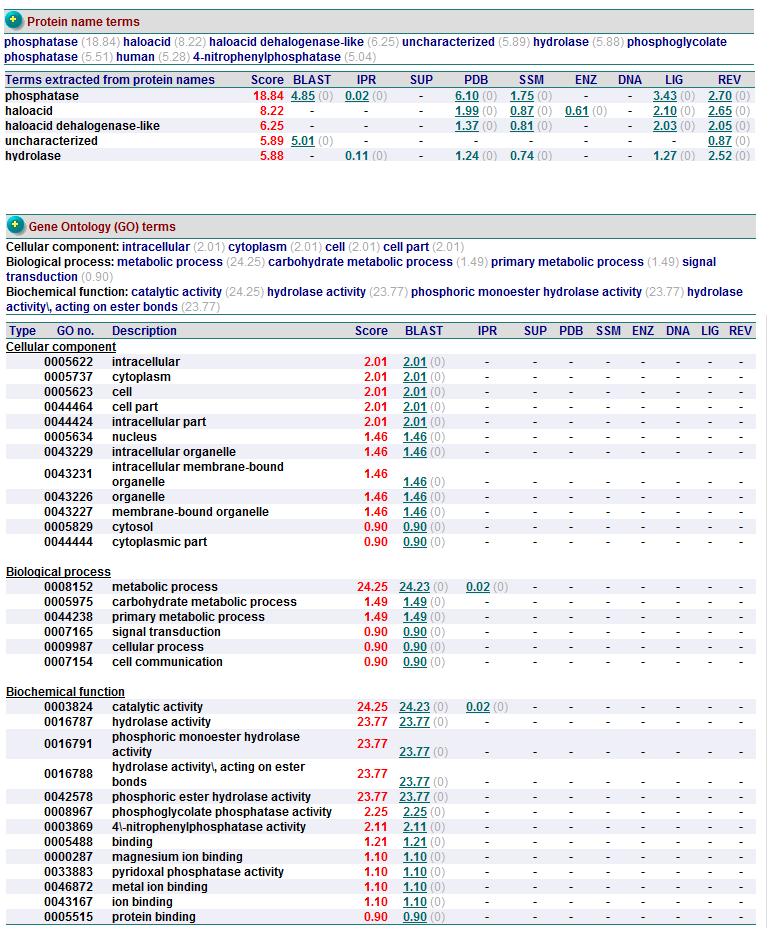
Function