Ssu72 Results: Difference between revisions
| Line 13: | Line 13: | ||
===Literature review and ProFunc results=== | ===Literature review and ProFunc results=== | ||
The human Ssu72 protein, for which there exist a number of eukaryotic homologues (Sun and Hampsey 1996), was first proposed to function as a protein tyrosine phosphatase (Meinhart, Silberzahn, & Cramer, 2003). Therefore, a ProFunc analysis was carried out on our candidate protein, Drosophila Ssu72, with the purpose of comparing components of its primary and secondary structure with proteins that do not necessarily have a high level of overall sequence similarity and thus would be unlikely to surface on a non-targeted search of a large database (ie.by BLAST). Despite offering a number of powerful results (see Figures 5 - 9), the ProFunc analysis of the Drosophila Ssu72 protein returned just two sets of informative data. The first of these was from the SSM (Secondary Structure Matching) program, which 'identifies proteins in the PDB that have a similar fold to that of the query structure' (Krissinel & Henrick, 2004). The summary of the results of this analysis (see Figures 10 and 11) is by itself promising, as the program exclusively returned proteins which function as phosphatases, all but one of which specifically has phosphotyrosine as a substrate. | The human Ssu72 protein, for which there exist a number of eukaryotic homologues (Sun and Hampsey 1996), was first proposed to function as a protein tyrosine phosphatase (Meinhart, Silberzahn, & Cramer, 2003). Therefore, a ProFunc analysis was carried out on our candidate protein, Drosophila Ssu72, with the purpose of comparing components of its primary and secondary structure with proteins that do not necessarily have a high level of overall sequence similarity and thus would be unlikely to surface on a non-targeted search of a large database (ie.by BLAST). Despite offering a number of powerful results (see Figures 5 - 9), the ProFunc analysis of the Drosophila Ssu72 protein returned just two sets of informative data. The first of these was from the SSM (Secondary Structure Matching) program, which 'identifies proteins in the PDB that have a similar fold to that of the query structure' (Krissinel & Henrick, 2004). The summary of the results of this analysis (see Figures 10 and 11) is by itself promising, as the program exclusively returned proteins which function as phosphatases, all but one of which specifically has phosphotyrosine as a substrate. | ||
In the next set of results, the so-called 'reverse template' program, the '[target] structure ... is broken up into a large number of templates and these in turn are scanned against a representative set of structures in the PDB' (Laskowski, Watson & Thornton, 2005). In effect, the queried sequence is compared against a database of known catalytic sites for matches. The results (see Figures ), show that, while there is only a low level of overall sequence identity (~ 20%) between the target structure and the hits returned by the program, the Drosophila Ssu72 protein shares three important residues - Val12(A), Cys13(A), Ser20(A) - with a number of different protein tyrosine phosphatases (see Figures ) | |||
demonstrate that, while there is only a low level of overall sequence identity (~ 20%) between the target structure and the hits returned by the program, the Drosophila Ssu72 protein shares three important residues, | |||
| Line 23: | Line 28: | ||
image:Figure_10.png|'''Figure 10''': 10 structural matches returned from the Secondary Structure Matching program. | image:Figure_10.png|'''Figure 10''': 10 structural matches returned from the Secondary Structure Matching program. | ||
image:Figure_11.png|'''Figure 11''': Alignment of secondary structure schematics from the 10 structural matches returned from the SSM program. | image:Figure_11.png|'''Figure 11''': Alignment of secondary structure schematics from the 10 structural matches returned from the SSM program. | ||
image:Figure_12.png|'''Figure 12''': Reverse template results - certain matches. | |||
image:Figure_13.png|'''Figure 13''': Reverse template results - probable matches | |||
</gallery></center> | </gallery></center> | ||
Laskowski RA, Watson JD and Thornton JM (2005). Protein function prediction using local 3D templates. J. Mol. Biol., 351, 614-626. | |||
===References=== | ===References=== | ||
Revision as of 14:14, 11 June 2009
Function
BLAST results
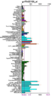
Running a protein BLAST search on 'serine phosphatase of RNA polymerase II CTD (SSU72 superfamily)' from Drosophila melenogaster (the 'Drosophila Ssu72 protein') returned a large number of highly relevant (bit score > 200) sequences that were of a very similar length (see Figure 1) to that of the query. However, the first 27 of those sequences were not considered relevant for the purposes of this paper for a number of reasons. For instance, some were merely homologous proteins in other closely related Drosophila species that had been identified in a similarly automated manner to this one and for which there were no PubMed publications available and thus no information that could be used to easily infer functionally relevant details. Similarly, other proteins, in more distantly related organisms, had not been functionally characterised by experimentation or structural analysis, presumably because they had been identified by high-throughput sequencing and had seen minimal, if any, human input. Some BLAST hits were labelled as hypothetical proteins and were therefore not necessarily protein coding and biologically expressed. Notwithstanding this, the first relevant protein identified was 'SSU72 RNA polymerase II CTD phosphatase homolog (S. cerevisiae)', the human Ssu72 protein, for which there were a small number of articles available on PubMed (see Figure 2), as well as expression data from SymAtlas (see Figure 3). This protein was also found to have a 60.21% sequence identity with the Drosophila Ssu72 protein, with no gaps in the alignment. Thus, a review of the literature surrounding it was considered a suitable entry-point in the process of this analysis.
Literature review and ProFunc results
The human Ssu72 protein, for which there exist a number of eukaryotic homologues (Sun and Hampsey 1996), was first proposed to function as a protein tyrosine phosphatase (Meinhart, Silberzahn, & Cramer, 2003). Therefore, a ProFunc analysis was carried out on our candidate protein, Drosophila Ssu72, with the purpose of comparing components of its primary and secondary structure with proteins that do not necessarily have a high level of overall sequence similarity and thus would be unlikely to surface on a non-targeted search of a large database (ie.by BLAST). Despite offering a number of powerful results (see Figures 5 - 9), the ProFunc analysis of the Drosophila Ssu72 protein returned just two sets of informative data. The first of these was from the SSM (Secondary Structure Matching) program, which 'identifies proteins in the PDB that have a similar fold to that of the query structure' (Krissinel & Henrick, 2004). The summary of the results of this analysis (see Figures 10 and 11) is by itself promising, as the program exclusively returned proteins which function as phosphatases, all but one of which specifically has phosphotyrosine as a substrate.
In the next set of results, the so-called 'reverse template' program, the '[target] structure ... is broken up into a large number of templates and these in turn are scanned against a representative set of structures in the PDB' (Laskowski, Watson & Thornton, 2005). In effect, the queried sequence is compared against a database of known catalytic sites for matches. The results (see Figures ), show that, while there is only a low level of overall sequence identity (~ 20%) between the target structure and the hits returned by the program, the Drosophila Ssu72 protein shares three important residues - Val12(A), Cys13(A), Ser20(A) - with a number of different protein tyrosine phosphatases (see Figures )
demonstrate that, while there is only a low level of overall sequence identity (~ 20%) between the target structure and the hits returned by the program, the Drosophila Ssu72 protein shares three important residues,
Laskowski RA, Watson JD and Thornton JM (2005). Protein function prediction using local 3D templates. J. Mol. Biol., 351, 614-626.
References
Krissinel E and Henrick K (2004). Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions Acta Cryst., D60, 2256-2268.
Z.-W. Sun and M. Hampsey, Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential gene encoding a novel protein that affects transcription start site selection in vivo, Mol. Cell. Biol. 16 (1996), pp. 1557–1566.
Evolution
Heading
Structure
Heading
Abstract | Introduction | Results | Discussion | Conclusion | Method | References